Structure determination of an amorphous drug through large-scale NMR predictions
- PMID: 34016980
- PMCID: PMC8137699
- DOI: 10.1038/s41467-021-23208-7
Structure determination of an amorphous drug through large-scale NMR predictions
Abstract
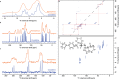
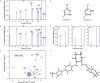
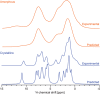
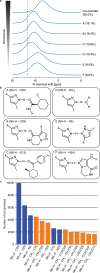
Knowledge of the structure of amorphous solids can direct, for example, the optimization of pharmaceutical formulations, but atomic-level structure determination in amorphous molecular solids has so far not been possible. Solid-state nuclear magnetic resonance (NMR) is among the most popular methods to characterize amorphous materials, and molecular dynamics (MD) simulations can help describe the structure of disordered materials. However, directly relating MD to NMR experiments in molecular solids has been out of reach until now because of the large size of these simulations. Here, using a machine learning model of chemical shifts, we determine the atomic-level structure of the hydrated amorphous drug AZD5718 by combining dynamic nuclear polarization-enhanced solid-state NMR experiments with predicted chemical shifts for MD simulations of large systems. From these amorphous structures we then identify H-bonding motifs and relate them to local intermolecular complex formation energies.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hughes CE, Boughdiri I, Bouakkaz C, Williams PA, Harris KDM. Elucidating the crystal structure of dl-arginine by combined powder X-ray diffraction data analysis and periodic DFT-D calculations. Cryst. Growth Des. 2017;18:42–46. doi: 10.1021/acs.cgd.7b01412. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

