The impact of chromosomal fusions on 3D genome folding and recombination in the germ line
- PMID: 34016985
- PMCID: PMC8137915
- DOI: 10.1038/s41467-021-23270-1
The impact of chromosomal fusions on 3D genome folding and recombination in the germ line
Abstract
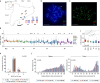
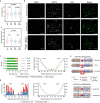
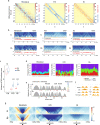
The spatial folding of chromosomes inside the nucleus has regulatory effects on gene expression, yet the impact of genome reshuffling on this organization remains unclear. Here, we take advantage of chromosome conformation capture in combination with single-nucleotide polymorphism (SNP) genotyping and analysis of crossover events to study how the higher-order chromatin organization and recombination landscapes are affected by chromosomal fusions in the mammalian germ line. We demonstrate that chromosomal fusions alter the nuclear architecture during meiosis, including an increased rate of heterologous interactions in primary spermatocytes, and alterations in both chromosome synapsis and axis length. These disturbances in topology were associated with changes in genomic landscapes of recombination, resulting in detectable genomic footprints. Overall, we show that chromosomal fusions impact the dynamic genome topology of germ cells in two ways: (i) altering chromosomal nuclear occupancy and synapsis, and (ii) reshaping landscapes of recombination.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Chromosome synapsis and recombination in simple and complex chromosomal heterozygotes of tuco-tuco (Ctenomys talarum: Rodentia: Ctenomyidae).Chromosome Res. 2014 Sep;22(3):351-63. doi: 10.1007/s10577-014-9429-5. Epub 2014 Jun 13. Chromosome Res. 2014. PMID: 24924853
-
Multiple Genomic Landscapes of Recombination and Genomic Divergence in Wild Populations of House Mice-The Role of Chromosomal Fusions and Prdm9.Mol Biol Evol. 2024 Apr 2;41(4):msae063. doi: 10.1093/molbev/msae063. Mol Biol Evol. 2024. PMID: 38513632 Free PMC article.
-
H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis.Dev Cell. 2003 Apr;4(4):497-508. doi: 10.1016/s1534-5807(03)00093-5. Dev Cell. 2003. PMID: 12689589
-
Homologue pairing, recombination and segregation in Caenorhabditis elegans.Genome Dyn. 2009;5:43-55. doi: 10.1159/000166618. Genome Dyn. 2009. PMID: 18948706 Review.
-
Chromosome territories, nuclear architecture and gene regulation in mammalian cells.Nat Rev Genet. 2001 Apr;2(4):292-301. doi: 10.1038/35066075. Nat Rev Genet. 2001. PMID: 11283701 Review.
Cited by
-
Strategies for meiotic sex chromosome dynamics and telomeric elongation in Marsupials.PLoS Genet. 2022 Feb 7;18(2):e1010040. doi: 10.1371/journal.pgen.1010040. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35130272 Free PMC article.
-
Celebrating Mendel, McClintock, and Darlington: On end-to-end chromosome fusions and nested chromosome fusions.Plant Cell. 2022 Jul 4;34(7):2475-2491. doi: 10.1093/plcell/koac116. Plant Cell. 2022. PMID: 35441689 Free PMC article. Review.
-
Nascent evolution of recombination rate differences as a consequence of chromosomal rearrangements.PLoS Genet. 2023 Aug 7;19(8):e1010717. doi: 10.1371/journal.pgen.1010717. eCollection 2023 Aug. PLoS Genet. 2023. PMID: 37549188 Free PMC article.
-
Recombination plasticity in response to temperature variation in reptiles.PLoS Genet. 2025 Aug 4;21(8):e1011772. doi: 10.1371/journal.pgen.1011772. eCollection 2025 Aug. PLoS Genet. 2025. PMID: 40758762 Free PMC article.
-
Chromosome Changes in Soma and Germ Line: Heritability and Evolutionary Outcome.Genes (Basel). 2022 Mar 28;13(4):602. doi: 10.3390/genes13040602. Genes (Basel). 2022. PMID: 35456408 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

