The expanding field of non-canonical RNA capping: new enzymes and mechanisms
- PMID: 34017598
- PMCID: PMC8131947
- DOI: 10.1098/rsos.201979
The expanding field of non-canonical RNA capping: new enzymes and mechanisms
Abstract
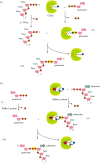
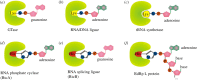
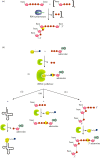
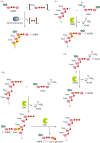
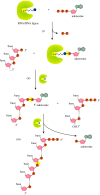
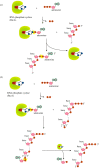
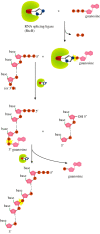
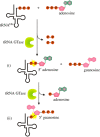
Recent years witnessed the discovery of ubiquitous and diverse 5'-end RNA cap-like modifications in prokaryotes as well as in eukaryotes. These non-canonical caps include metabolic cofactors, such as NAD+/NADH, FAD, cell wall precursors UDP-GlcNAc, alarmones, e.g. dinucleotides polyphosphates, ADP-ribose and potentially other nucleoside derivatives. They are installed at the 5' position of RNA via template-dependent incorporation of nucleotide analogues as an initiation substrate by RNA polymerases. However, the discovery of NAD-capped processed RNAs in human cells suggests the existence of alternative post-transcriptional NC capping pathways. In this review, we compiled growing evidence for a number of these other mechanisms which produce various non-canonically capped RNAs and a growing repertoire of capping small molecules. Enzymes shown to be involved are ADP-ribose polymerases, glycohydrolases and tRNA synthetases, and may potentially include RNA 3'-phosphate cyclases, tRNA guanylyl transferases, RNA ligases and ribozymes. An emerging rich variety of capping molecules and enzymes suggests an unrecognized level of complexity of RNA metabolism.
Keywords: RNA processing; capping; non-canonical capping.
© 2021 The Authors.
Figures

Similar articles
-
Preparation of RNAs with non-canonical 5' ends using novel di- and trinucleotide reagents for co-transcriptional capping.Front Mol Biosci. 2022 Aug 19;9:854170. doi: 10.3389/fmolb.2022.854170. eCollection 2022. Front Mol Biosci. 2022. PMID: 36060251 Free PMC article.
-
Noncanonical RNA-capping: Discovery, mechanism, and physiological role debate.Wiley Interdiscip Rev RNA. 2019 Mar;10(2):e1512. doi: 10.1002/wrna.1512. Epub 2018 Oct 23. Wiley Interdiscip Rev RNA. 2019. PMID: 30353673 Review.
-
If the 5' cap fits (wear it) - Non-canonical RNA capping.RNA Biol. 2024 Jan;21(1):1-13. doi: 10.1080/15476286.2024.2372138. Epub 2024 Jul 15. RNA Biol. 2024. PMID: 39007883 Free PMC article. Review.
-
RNA capping by mitochondrial and multi-subunit RNA polymerases.Transcription. 2018;9(5):292-297. doi: 10.1080/21541264.2018.1456258. Epub 2018 Apr 25. Transcription. 2018. PMID: 29624107 Free PMC article. Review.
-
Identification and in vitro characterization of UDP-GlcNAc-RNA cap-modifying and decapping enzymes.Nucleic Acids Res. 2024 Jun 10;52(10):5438-5450. doi: 10.1093/nar/gkae353. Nucleic Acids Res. 2024. PMID: 38716860 Free PMC article.
Cited by
-
RNA modifications and their role in gene expression.Front Mol Biosci. 2025 Apr 25;12:1537861. doi: 10.3389/fmolb.2025.1537861. eCollection 2025. Front Mol Biosci. 2025. PMID: 40351534 Free PMC article. Review.
-
PAR-dCLIP: Enabling detection of RNA binding protein target transcripts bound at 5' termini through the incorporation of a decapping step.Methods Enzymol. 2024;705:159-222. doi: 10.1016/bs.mie.2024.08.003. Epub 2024 Sep 7. Methods Enzymol. 2024. PMID: 39389663 Free PMC article.
-
Application of Mammalian Nudix Enzymes to Capped RNA Analysis.Pharmaceuticals (Basel). 2024 Sep 11;17(9):1195. doi: 10.3390/ph17091195. Pharmaceuticals (Basel). 2024. PMID: 39338357 Free PMC article. Review.
-
Preparation of RNAs with non-canonical 5' ends using novel di- and trinucleotide reagents for co-transcriptional capping.Front Mol Biosci. 2022 Aug 19;9:854170. doi: 10.3389/fmolb.2022.854170. eCollection 2022. Front Mol Biosci. 2022. PMID: 36060251 Free PMC article.
-
RNA biology takes root in plant systems.Plant Direct. 2022 Sep 6;6(9):e445. doi: 10.1002/pld3.445. eCollection 2022 Sep. Plant Direct. 2022. PMID: 36091875 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

