CNVxplorer: a web tool to assist clinical interpretation of CNVs in rare disease patients
- PMID: 34019647
- PMCID: PMC8262689
- DOI: 10.1093/nar/gkab347
CNVxplorer: a web tool to assist clinical interpretation of CNVs in rare disease patients
Abstract
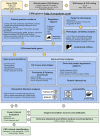
Copy Number Variants (CNVs) are an important cause of rare diseases. Array-based Comparative Genomic Hybridization tests yield a ∼12% diagnostic rate, with ∼8% of patients presenting CNVs of unknown significance. CNVs interpretation is particularly challenging on genomic regions outside of those overlapping with previously reported structural variants or disease-associated genes. Recent studies showed that a more comprehensive evaluation of CNV features, leveraging both coding and non-coding impacts, can significantly improve diagnostic rates. However, currently available CNV interpretation tools are mostly gene-centric or provide only non-interactive annotations difficult to assess in the clinical practice. Here, we present CNVxplorer, a web server suited for the functional assessment of CNVs in a clinical diagnostic setting. CNVxplorer mines a comprehensive set of clinical, genomic, and epigenomic features associated with CNVs. It provides sequence constraint metrics, impact on regulatory elements and topologically associating domains, as well as expression patterns. Analyses offered cover (a) agreement with patient phenotypes; (b) visualizations of associations among genes, regulatory elements and transcription factors; (c) enrichment on functional and pathway annotations and (d) co-occurrence of terms across PubMed publications related to the query CNVs. A flexible evaluation workflow allows dynamic re-interrogation in clinical sessions. CNVxplorer is publicly available at http://cnvxplorer.com.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Weischenfeldt J., Symmons O., Spitz F., Korbel J.O.. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013; 14:125–138. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

