Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease
- PMID: 34019811
- PMCID: PMC8221567
- DOI: 10.1016/j.molcel.2021.04.028
Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease
Abstract
ARH3/ADPRHL2 and PARG are the primary enzymes reversing ADP-ribosylation in vertebrates, yet their functions in vivo remain unclear. ARH3 is the only hydrolase able to remove serine-linked mono(ADP-ribose) (MAR) but is much less efficient than PARG against poly(ADP-ribose) (PAR) chains in vitro. Here, by using ARH3-deficient cells, we demonstrate that endogenous MARylation persists on chromatin throughout the cell cycle, including mitosis, and is surprisingly well tolerated. Conversely, persistent PARylation is highly toxic and has distinct physiological effects, in particular on active transcription histone marks such as H3K9ac and H3K27ac. Furthermore, we reveal a synthetic lethal interaction between ARH3 and PARG and identify loss of ARH3 as a mechanism of PARP inhibitor resistance, both of which can be exploited in cancer therapy. Finally, we extend our findings to neurodegeneration, suggesting that patients with inherited ARH3 deficiency suffer from stress-induced pathogenic increase in PARylation that can be mitigated by PARP inhibition.
Keywords: ADP-ribosylation; ARH3/ADPRHL2; BRCA; DNA damage; PARG; PARP inhibitor; cancer; chromatin; neurodegeneration; telomere.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
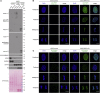


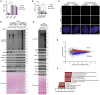
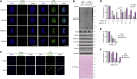

References
-
- Amé J.-C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., de Murcia G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0400653/MRC_/Medical Research Council/United Kingdom
- BB/R016836/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P30 CA047904/CA/NCI NIH HHS/United States
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 16304/CRUK_/Cancer Research UK/United Kingdom
- 210634/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- BB/R007195/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C35050/A22284/CRUK_/Cancer Research UK/United Kingdom
- 22284/CRUK_/Cancer Research UK/United Kingdom
- 210634 /WT_/Wellcome Trust/United Kingdom
- 210641/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA207209/CA/NCI NIH HHS/United States
- 101794 /WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

