Orchard Management and Landscape Context Mediate the Pear Floral Microbiome
- PMID: 34020936
- PMCID: PMC8276804
- DOI: 10.1128/AEM.00048-21
Orchard Management and Landscape Context Mediate the Pear Floral Microbiome
Abstract
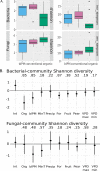
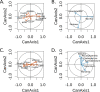
Crop-associated microbiota are a key factor affecting host health and productivity. Most crops are grown within heterogeneous landscapes, and interactions between management practices and landscape context often affect plant and animal biodiversity in agroecosystems. However, whether these same factors typically affect crop-associated microbiota is less clear. Here, we assessed whether orchard management strategies and landscape context affected bacterial and fungal communities in pear (Pyrus communis) flowers. We found that bacteria and fungi responded differently to management schemes. Organically certified orchards had higher fungal diversity in flowers than conventional or bio-based integrated pest management (IPM) orchards, but organic orchards had the lowest bacterial diversity. Orchard management scheme also best predicted the distribution of several important bacterial and fungal genera that either cause or suppress disease; organic and bio-based IPM best explained the distributions of bacterial and fungal genera, respectively. Moreover, patterns of bacterial and fungal diversity were affected by interactions between management, landscape context, and climate. When examining the similarity of bacterial and fungal communities across sites, both abundance- and taxon-related turnovers were mediated primarily by orchard management scheme and landscape context and, specifically, the amount of land in cultivation. Our study reveals local- and landscape-level drivers of floral microbiome structure in a major fruit crop, providing insights that can inform microbiome management to promote host health and high-yielding quality fruit. IMPORTANCE Proper crop management during bloom is essential for producing disease-free tree fruit. Tree fruits are often grown in heterogeneous landscapes; however, few studies have assessed whether landscape context and crop management affect the floral microbiome, which plays a critical role in shaping plant health and disease tolerance. Such work is key for identification of tactics and/or contexts where beneficial microbes proliferate and pathogenic microbes are limited. Here, we characterize the floral microbiome of pear crops in Washington State, where major production occurs in intermountain valleys and basins with variable elevation and microclimates. Our results show that both local-level (crop management) and landscape-level (habitat types and climate) factors affect floral microbiota but in disparate ways for each kingdom. More broadly, these findings can potentially inform microbiome management in orchards for promotion of host health and high-quality yields.
Keywords: Pyrus communis; flower microbiome; integrated pest management; landscape heterogeneity.
Figures



References
-
- Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C. 2015. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol Fertil Soils 51:403–415. 10.1007/s00374-015-0996-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

