IL-6 effector function of group 2 innate lymphoid cells (ILC2) is NOD2 dependent
- PMID: 34021026
- PMCID: PMC7611333
- DOI: 10.1126/sciimmunol.abe5084
IL-6 effector function of group 2 innate lymphoid cells (ILC2) is NOD2 dependent
Abstract
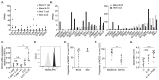
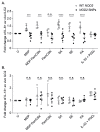
Cutaneous group 2 innate lymphoid cells (ILC2) are spatially and epigenetically poised to respond to barrier compromise and associated immunological threats. ILC2, lacking rearranged antigen-specific receptors, are primarily activated by damage-associated cytokines and respond with type 2 cytokine production. To investigate ILC2 potential for direct sensing of skin pathogens and allergens, we performed RNA sequencing of ILC2 derived from in vivo challenged human skin or blood. We detected expression of NOD2 and TLR2 by skin and blood ILC2. Stimulation of ILC2 with TLR2 agonist alone not only induced interleukin-5 (IL-5) and IL-13 expression but also elicited IL-6 expression in combination with Staphylococcus aureus muramyl dipeptide (MDP). Heat-killed skin-resident bacteria provoked an IL-6 profile in ILC2 in vitro that was notably impaired in ILC2 derived from patients with nucleotide-binding oligomerization domain-containing protein 2 (NOD2) mutations. In addition, we show that NOD2 signaling can stimulate autophagy in ILC2, which was also impaired in patients with NOD2 mutations. Here, we have identified a role for ILC2 NOD2 signaling in the differential regulation of ILC2-derived IL-6 and have reported a previously unrecognized pathway of direct ILC2 bacterial sensing.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





References
-
- Cherrier DE, Serafini N, Di Santo JP. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity. 2018;48:1091–1103. - PubMed
-
- Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U105178805/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/5/MRC_/Medical Research Council/United Kingdom
- MC_PC_MR/S025952/1/MRC_/Medical Research Council/United Kingdom
- MC_U105178805/MRC_/Medical Research Council/United Kingdom
- MR/K018779/1/MRC_/Medical Research Council/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- MR/S036377/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/7/MRC_/Medical Research Council/United Kingdom
- MC_UU_12010/7/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- CF7720/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

