RNA helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis B virus replicating hepatocytes
- PMID: 34021034
- PMCID: PMC8606016
- DOI: 10.1136/gutjnl-2020-323126
RNA helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis B virus replicating hepatocytes
Abstract
Objective: RNA helicase DDX5 is downregulated during HBV replication and poor prognosis HBV-related hepatocellular carcinoma (HCC). The objective of this study is to investigate the role of DDX5 in interferon (IFN) signalling. We provide evidence of a novel mechanism involving DDX5 that enables translation of transcription factor STAT1 mediating the IFN response.
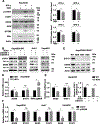
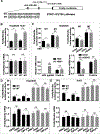
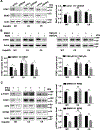
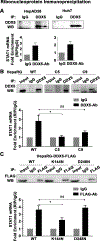
Design and results: Molecular, pharmacological and biophysical assays were used together with cellular models of HBV replication, HCC cell lines and liver tumours. We demonstrate that DDX5 regulates STAT1 mRNA translation by resolving a G-quadruplex (rG4) RNA structure, proximal to the 5' end of STAT1 5'UTR. We employed luciferase reporter assays comparing wild type (WT) versus mutant rG4 sequence, rG4-stabilising compounds, CRISPR/Cas9 editing of the STAT1-rG4 sequence and circular dichroism determination of the rG4 structure. STAT1-rG4 edited cell lines were resistant to the effect of rG4-stabilising compounds in response to IFN-α, while HCC cell lines expressing low DDX5 exhibited reduced IFN response. Ribonucleoprotein and electrophoretic mobility assays demonstrated direct and selective binding of RNA helicase-active DDX5 to the WT STAT1-rG4 sequence. Immunohistochemistry of normal liver and liver tumours demonstrated that absence of DDX5 corresponded to absence of STAT1. Significantly, knockdown of DDX5 in HBV infected HepaRG cells reduced the anti-viral effect of IFN-α.
Conclusion: RNA helicase DDX5 resolves a G-quadruplex structure in 5'UTR of STAT1 mRNA, enabling STAT1 translation. We propose that DDX5 is a key regulator of the dynamic range of IFN response during innate immunity and adjuvant IFN-α therapy.
Keywords: hepatitis B; hepatocellular carcinoma; hepatocyte; interferon-alpha; molecular mechanisms.
© Author(s) (or their employer(s)) 2022. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
-
- Chen K, Liu J, Liu S, et al. Methyltransferase SETD2-Mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 2017;170:492–506. - PubMed
-
- Liu S, Jiang M, Wang W, et al. Nuclear RNF2 inhibits interferon function by promoting K33-linked STAT1 disassociation from DNA. Nat Immunol 2018;19:41–52. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
