Cytoplasmic FUS triggers early behavioral alterations linked to cortical neuronal hyperactivity and inhibitory synaptic defects
- PMID: 34021132
- PMCID: PMC8140148
- DOI: 10.1038/s41467-021-23187-9
Cytoplasmic FUS triggers early behavioral alterations linked to cortical neuronal hyperactivity and inhibitory synaptic defects
Abstract
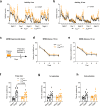
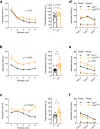
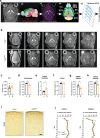
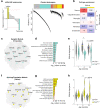
Gene mutations causing cytoplasmic mislocalization of the RNA-binding protein FUS lead to severe forms of amyotrophic lateral sclerosis (ALS). Cytoplasmic accumulation of FUS is also observed in other diseases, with unknown consequences. Here, we show that cytoplasmic mislocalization of FUS drives behavioral abnormalities in knock-in mice, including locomotor hyperactivity and alterations in social interactions, in the absence of widespread neuronal loss. Mechanistically, we identified a progressive increase in neuronal activity in the frontal cortex of Fus knock-in mice in vivo, associated with altered synaptic gene expression. Synaptic ultrastructural and morphological defects were more pronounced in inhibitory than excitatory synapses and associated with increased synaptosomal levels of FUS and its RNA targets. Thus, cytoplasmic FUS triggers synaptic deficits, which is leading to increased neuronal activity in frontal cortex and causing related behavioral phenotypes. These results indicate that FUS mislocalization may trigger deleterious phenotypes beyond motor neuron impairment in ALS, likely relevant also for other neurodegenerative diseases characterized by FUS mislocalization.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

