Co-development of central and peripheral neurons with trunk mesendoderm in human elongating multi-lineage organized gastruloids
- PMID: 34021144
- PMCID: PMC8140076
- DOI: 10.1038/s41467-021-23294-7
Co-development of central and peripheral neurons with trunk mesendoderm in human elongating multi-lineage organized gastruloids
Abstract
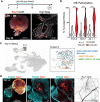
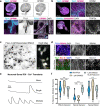
Stem cell technologies including self-assembling 3D tissue models provide access to early human neurodevelopment and fundamental insights into neuropathologies. Gastruloid models have not been used to investigate co-developing central and peripheral neuronal systems with trunk mesendoderm which we achieve here in elongating multi-lineage organized (EMLO) gastruloids. We evaluate EMLOs over a forty-day period, applying immunofluorescence of multi-lineage and functional biomarkers, including day 16 single-cell RNA-Seq, and evaluation of ectodermal and non-ectodermal neural crest cells (NCCs). We identify NCCs that differentiate to form peripheral neurons integrated with an upstream spinal cord region after day 8. This follows initial EMLO polarization events that coordinate with endoderm differentiation and primitive gut tube formation during multicellular spatial reorganization. This combined human central-peripheral nervous system model of early organogenesis highlights developmental events of mesendoderm and neuromuscular trunk regions and enables systemic studies of tissue interactions and innervation of neuromuscular, enteric and cardiac relevance.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

