RTEL1 influences the abundance and localization of TERRA RNA
- PMID: 34021146
- PMCID: PMC8140157
- DOI: 10.1038/s41467-021-23299-2
RTEL1 influences the abundance and localization of TERRA RNA
Abstract
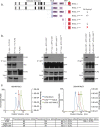
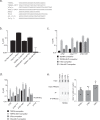
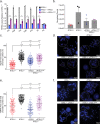
Telomere repeat containing RNAs (TERRAs) are a family of long non-coding RNAs transcribed from the subtelomeric regions of eukaryotic chromosomes. TERRA transcripts can form R-loops at chromosome ends; however the importance of these structures or the regulation of TERRA expression and retention in telomeric R-loops remain unclear. Here, we show that the RTEL1 (Regulator of Telomere Length 1) helicase influences the abundance and localization of TERRA in human cells. Depletion of RTEL1 leads to increased levels of TERRA RNA while reducing TERRA-containing R loops at telomeres. In vitro, RTEL1 shows a strong preference for binding G-quadruplex structures which form in TERRA. This binding is mediated by the C-terminal region of RTEL1, and is independent of the RTEL1 helicase domain. RTEL1 binding to TERRA appears to be essential for cell viability, underscoring the importance of this function. Degradation of TERRA-containing R-loops by overexpression of RNAse H1 partially recapitulates the increased TERRA levels and telomeric instability associated with RTEL1 deficiency. Collectively, these data suggest that regulation of TERRA is a key function of the RTEL1 helicase, and that loss of that function may contribute to the disease phenotypes of patients with RTEL1 mutations.
Conflict of interest statement
J.H.J.P. is a consultant for Novus Biologicals and ATROPOS Therapeutics, which are not competing interests with this study. The remaining authors declare no competing interests.
Figures



Similar articles
-
Rapid dynamics allow the low-abundance RTEL1 helicase to promote telomere replication.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf177. doi: 10.1093/nar/gkaf177. Nucleic Acids Res. 2025. PMID: 40087886 Free PMC article.
-
RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops.Nature. 2020 Nov;587(7833):303-308. doi: 10.1038/s41586-020-2815-6. Epub 2020 Oct 14. Nature. 2020. PMID: 33057192 Free PMC article.
-
Structural and biochemical characterization of the C-terminal region of the human RTEL1 helicase.Protein Sci. 2024 Sep;33(9):e5093. doi: 10.1002/pro.5093. Protein Sci. 2024. PMID: 39180489 Free PMC article.
-
Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity.Front Genet. 2015 Apr 14;6:143. doi: 10.3389/fgene.2015.00143. eCollection 2015. Front Genet. 2015. PMID: 25926849 Free PMC article. Review.
-
The many faces of the helicase RTEL1 at telomeres and beyond.Trends Cell Biol. 2024 Feb;34(2):109-121. doi: 10.1016/j.tcb.2023.07.002. Epub 2023 Jul 31. Trends Cell Biol. 2024. PMID: 37532653 Review.
Cited by
-
Telomere Shortening and Its Association with Cell Dysfunction in Lung Diseases.Int J Mol Sci. 2021 Dec 31;23(1):425. doi: 10.3390/ijms23010425. Int J Mol Sci. 2021. PMID: 35008850 Free PMC article. Review.
-
A Large-Scale Exome-Wide Association Study Identifies Novel Germline Mutations in Lung Cancer.Am J Respir Crit Care Med. 2023 Aug 1;208(3):280-289. doi: 10.1164/rccm.202212-2199OC. Am J Respir Crit Care Med. 2023. PMID: 37167549 Free PMC article.
-
Defective X-chromosome inactivation and cancer risk in women.Commun Biol. 2025 Feb 22;8(1):289. doi: 10.1038/s42003-025-07691-y. Commun Biol. 2025. PMID: 39987288 Free PMC article.
-
Rapid dynamics allow the low-abundance RTEL1 helicase to promote telomere replication.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf177. doi: 10.1093/nar/gkaf177. Nucleic Acids Res. 2025. PMID: 40087886 Free PMC article.
-
Maternal methionine supplementation during gestation alters alternative splicing and DNA methylation in bovine skeletal muscle.BMC Genomics. 2021 Oct 30;22(1):780. doi: 10.1186/s12864-021-08065-4. BMC Genomics. 2021. PMID: 34717556 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

