Complete mitochondrial genomes and phylogenetic relationships of the genera Nephila and Trichonephila (Araneae, Araneoidea)
- PMID: 34021208
- PMCID: PMC8139964
- DOI: 10.1038/s41598-021-90162-1
Complete mitochondrial genomes and phylogenetic relationships of the genera Nephila and Trichonephila (Araneae, Araneoidea)
Abstract
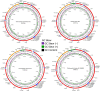
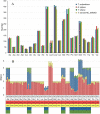
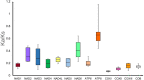
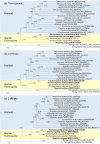
Spiders of the genera Nephila and Trichonephila are large orb-weaving spiders. In view of the lack of study on the mitogenome of these genera, and the conflicting systematic status, we sequenced (by next generation sequencing) and annotated the complete mitogenomes of N. pilipes, T. antipodiana and T. vitiana (previously N. vitiana) to determine their features and phylogenetic relationship. Most of the tRNAs have aberrant clover-leaf secondary structure. Based on 13 protein-coding genes (PCGs) and 15 mitochondrial genes (13 PCGs and two rRNA genes), Nephila and Trichonephila form a clade distinctly separated from the other araneid subfamilies/genera. T. antipodiana forms a lineage with T. vitiana in the subclade containing also T. clavata, while N. pilipes forms a sister clade to Trichonephila. The taxon vitiana is therefore a member of the genus Trichonephila and not Nephila as currently recognized. Studies on the mitogenomes of other Nephila and Trichonephila species and related taxa are needed to provide a potentially more robust phylogeny and systematics.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
A molecular phylogeny of nephilid spiders: evolutionary history of a model lineage.Mol Phylogenet Evol. 2013 Dec;69(3):961-79. doi: 10.1016/j.ympev.2013.06.008. Epub 2013 Jun 27. Mol Phylogenet Evol. 2013. PMID: 23811436
-
Characterization of the complete mitogenomes of two Neoscona spiders (Araneae: Araneidae) and its phylogenetic implications.Gene. 2016 Sep 30;590(2):298-306. doi: 10.1016/j.gene.2016.05.037. Epub 2016 May 31. Gene. 2016. PMID: 27259661
-
The complete mitochondrial genomes of two orb-weaving spider Cyrtarachne nagasakiensis (Strand, 1918) and Hypsosinga pygmaea (Sundevall, 1831) (Araneae: Araneidae).Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Jul;27(4):2811-2. doi: 10.3109/19401736.2015.1053079. Epub 2015 Jun 29. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26119123
-
Phylogenetic placement of the spider genus Nephila (Araneae: Araneoidea) inferred from rRNA and MaSp1 gene sequences.Zoolog Sci. 2004 Mar;21(3):343-51. doi: 10.2108/zsj.21.343. Zoolog Sci. 2004. PMID: 15056930
-
Systematics, phylogeny, and evolution of orb-weaving spiders.Annu Rev Entomol. 2014;59:487-512. doi: 10.1146/annurev-ento-011613-162046. Epub 2013 Oct 25. Annu Rev Entomol. 2014. PMID: 24160416 Review.
Cited by
-
New mitochondrial genomes of three whip spider species from the Amazon (Arachnida, Amblypygi) with phylogenetic relationships and comparative analysis.Sci Rep. 2024 Nov 1;14(1):26271. doi: 10.1038/s41598-024-77525-0. Sci Rep. 2024. PMID: 39487275 Free PMC article.
References
-
- World Spider Catalog (2020) Natural History Museum Bern. http://wsc.nmbe.ch. Accessed 25 July 2020
-
- Harvey MS, Austin AD, Adams M. The systematics and biology of the spider genus Nephila (Araneae: Nephilidae) in the Australasian region. Invertebr. Syst. 2007;21:407–451. doi: 10.1071/IS05016. - DOI
-
- Kuntner M. Phylogenetic systematics of the Gondwanan nephilid spider lineage Clitaetrinae (Araneae, Nephilidae) Zoolog. Scr. 2006;35:19–62. doi: 10.1111/j.1463-6409.2006.00220.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

