Tools for Assessing Cell-Cycle Progression in Plants
- PMID: 34021583
- PMCID: PMC8579159
- DOI: 10.1093/pcp/pcab066
Tools for Assessing Cell-Cycle Progression in Plants
Abstract
Estimation of cell-cycle parameters is crucial for understanding the developmental programs established during the formation of an organism. A number of complementary approaches have been developed and adapted to plants to assess the cell-cycle status in different proliferative tissues. The most classical methods relying on metabolic labeling are still very much employed and give valuable information on cell-cycle progression in fixed tissues. However, the growing knowledge of plant cell-cycle regulators with defined expression pattern together with the development of fluorescent proteins technology enabled the generation of fusion proteins that function individually or in conjunction as cell-cycle reporters. Together with the improvement of imaging techniques, in vivo live imaging to monitor plant cell-cycle progression in normal growth conditions or in response to different stimuli has been possible. Here, we review these tools and their specific outputs for plant cell-cycle analysis.
Keywords: Arabidopsis; Cell cycle; Live-imaging; Metabolic labeling; Reporter genes; plants.
© The Author(s) 2021. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
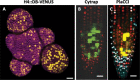
Figures

References
-
- Campilho A., Garcia B., Toorn H.V.D., Wijk H.V., Campilho A. and Scheres B. (2006) Time-lapse analysis of stem-cell divisions in the Arabidopsis thaliana root meristem. Plant J. 48: 619–627. - PubMed
-
- Caro E. and Gutierrez C. (2007) A green GEM: intriguing analogies with animal geminin. Trends Cell Biol. 17: 580–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

