Detection of SARS-CoV-2 genome and whole transcriptome sequencing in frontal cortex of COVID-19 patients
- PMID: 34022369
- PMCID: PMC8132498
- DOI: 10.1016/j.bbi.2021.05.012
Detection of SARS-CoV-2 genome and whole transcriptome sequencing in frontal cortex of COVID-19 patients
Abstract
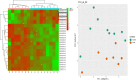
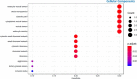
SARS-Cov-2 infection is frequently associated with Nervous System manifestations. However, it is not clear how SARS-CoV-2 can cause neurological dysfunctions and which molecular processes are affected in the brain. In this work, we examined the frontal cortex tissue of patients who died of COVID-19 for the presence of SARS-CoV-2, comparing qRT-PCR with ddPCR. We also investigated the transcriptomic profile of frontal cortex from COVID-19 patients and matched controls by RNA-seq analysis to characterize the transcriptional signature. Our data showed that SARS-CoV-2 could be detected by ddPCR in 8 (88%) of 9 examined samples while by qRT-PCR in one case only (11%). Transcriptomic analysis revealed that 11 genes (10 mRNAs and 1 lncRNA) were differential expressed when frontal cortex of COVID-19 patients were compared to controls. These genes fall into categories including hypoxia, hemoglobin-stabilizing protein, hydrogen peroxide processes. This work demonstrated that the quantity of viral RNA in frontal cortex is minimal and it can be detected only with a very sensitive method (ddPCR). Thus, it is likely that SARS-CoV-2 does not actively infect and replicate in the brain; its topography within encephalic structures remains uncertain. Moreover, COVID-19 may have a role on brain gene expression, since we observed an important downregulation of genes associated to hypoxia inducting factor system (HIF) that may inhibit the capacity of defense system during infection and oxigen deprivation, showing that hypoxia, well known multi organ condition associated to COVID-19, also marked the brain.
Keywords: Frontal cortex; Hemoglobin; Hypoxia; SARS-CoV-2; Transcriptome.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- K.S. Appelberg S. Gupta svensson akujsärvi, Sara & Ambikan, Anoop & Mikaeloff, Flora & Saccon, Elisa & Végvári, Akos & Benfeitas, Rui & Sperk, Maike & Ståhlberg, Marie & Krishnan, Shuba & Singh, Kamal & Penninger, Josef & Mirazimi, Ali & Neogi, Ujjwal. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerging Microbes & Infections. 9 2020 1 36 10.1080/22221751.2020.1799723. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

