Genome-wide identification, evolutionary estimation and functional characterization of two cotton CKI gene types
- PMID: 34022812
- PMCID: PMC8140429
- DOI: 10.1186/s12870-021-02990-y
Genome-wide identification, evolutionary estimation and functional characterization of two cotton CKI gene types
Abstract
Background: Casein kinase I (CKI) is a kind of serine/threonine protein kinase highly conserved in plants and animals. Although molecular function of individual member of CKI family has been investigated in Arabidopsis, little is known about their evolution and functions in Gossypium.
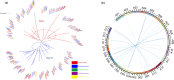
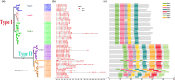
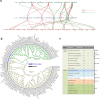
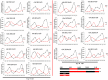
Results: In this study, five cotton species were applied to study CKI gene family in cotton, twenty-two species were applied to trace the origin and divergence of CKI genes. Four important insights were gained: (i) the cotton CKI genes were classified into two types based on their structural characteristics; (ii) two types of CKI genes expanded with tetraploid event in cotton; (iii) two types of CKI genes likely diverged about 1.5 billion years ago when red and green algae diverged; (iv) two types of cotton CKI genes which highly expressed in leaves showed stronger response to photoperiod (circadian clock) and light signal, and most two types of CKI genes highly expressed in anther showed identical heat inducible expression during anther development in tetraploid cotton (Gossypium hirsutum).
Conclusion: This study provides genome-wide insights into the evolutionary history of cotton CKI genes and lays a foundation for further investigation of the functional differentiation of two types of CKI genes in specific developmental processes and environmental stress conditions.
Keywords: Casein kinase I; Cotton; Evolutionary history; Gene expression.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genome-wide analysis and characterization of F-box gene family in Gossypium hirsutum L.BMC Genomics. 2019 Dec 19;20(1):993. doi: 10.1186/s12864-019-6280-2. BMC Genomics. 2019. PMID: 31856713 Free PMC article.
-
Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.).BMC Genet. 2017 Jun 12;18(1):54. doi: 10.1186/s12863-017-0517-3. BMC Genet. 2017. PMID: 28606097 Free PMC article.
-
Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum).BMC Genomics. 2014 Nov 6;15(1):961. doi: 10.1186/1471-2164-15-961. BMC Genomics. 2014. PMID: 25378022 Free PMC article.
-
Genome-wide analysis and comparison of the DNA-binding one zinc finger gene family in diploid and tetraploid cotton (Gossypium).PLoS One. 2020 Jun 29;15(6):e0235317. doi: 10.1371/journal.pone.0235317. eCollection 2020. PLoS One. 2020. PMID: 32598401 Free PMC article.
-
Genome-wide identification and characterization of LRR-RLKs reveal functional conservation of the SIF subfamily in cotton (Gossypium hirsutum).BMC Plant Biol. 2018 Sep 6;18(1):185. doi: 10.1186/s12870-018-1395-1. BMC Plant Biol. 2018. PMID: 30189845 Free PMC article.
Cited by
-
In vitro induction of tetraploids and their phenotypic and transcriptome analysis in Glehnia littoralis.BMC Plant Biol. 2024 May 22;24(1):439. doi: 10.1186/s12870-024-05154-w. BMC Plant Biol. 2024. PMID: 38778255 Free PMC article.
-
Unraveling the genetic and molecular basis of heat stress in cotton.Front Genet. 2024 Jun 11;15:1296622. doi: 10.3389/fgene.2024.1296622. eCollection 2024. Front Genet. 2024. PMID: 38919956 Free PMC article. Review.
-
Biological characterization and genomic profiling of a novel Providencia phage isolated from farm effluents.Arch Virol. 2025 Jul 29;170(8):187. doi: 10.1007/s00705-025-06373-8. Arch Virol. 2025. PMID: 40731076
References
-
- Tuazon PT, Traugh J. A casein kinase I and II–multipotential serine protein kinases: structure, function, and regulation. Adv Second Messenger Phosphoprotein Res. 1991;23:123–164. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

