A cyclic peptide inhibitor of the SARS-CoV-2 main protease
- PMID: 34023738
- PMCID: PMC8096527
- DOI: 10.1016/j.ejmech.2021.113530
A cyclic peptide inhibitor of the SARS-CoV-2 main protease
Abstract
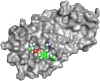
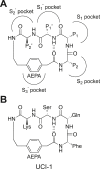
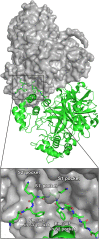
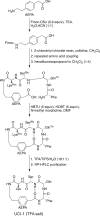
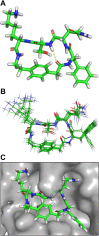
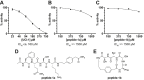
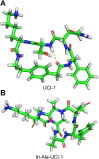
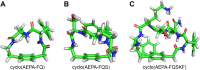
This paper presents the design and study of a first-in-class cyclic peptide inhibitor against the SARS-CoV-2 main protease (Mpro). The cyclic peptide inhibitor is designed to mimic the conformation of a substrate at a C-terminal autolytic cleavage site of Mpro. The cyclic peptide contains a [4-(2-aminoethyl)phenyl]-acetic acid (AEPA) linker that is designed to enforce a conformation that mimics a peptide substrate of Mpro. In vitro evaluation of the cyclic peptide inhibitor reveals that the inhibitor exhibits modest activity against Mpro and does not appear to be cleaved by the enzyme. Conformational searching predicts that the cyclic peptide inhibitor is fairly rigid, adopting a favorable conformation for binding to the active site of Mpro. Computational docking to the SARS-CoV-2 Mpro suggests that the cyclic peptide inhibitor can bind the active site of Mpro in the predicted manner. Molecular dynamics simulations provide further insights into how the cyclic peptide inhibitor may bind the active site of Mpro. Although the activity of the cyclic peptide inhibitor is modest, its design and study lays the groundwork for the development of additional cyclic peptide inhibitors against Mpro with improved activities.
Keywords: COVID-19; Cyclic peptide inhibitor; Cyclophane; Main protease; SARS-CoV-2.
Copyright © 2021 The Author(s). Published by Elsevier Masson SAS.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Structure-based drug design of an inhibitor of the SARS-CoV-2 (COVID-19) main protease using free software: A tutorial for students and scientists.Eur J Med Chem. 2021 Jun 5;218:113390. doi: 10.1016/j.ejmech.2021.113390. Epub 2021 Mar 20. Eur J Med Chem. 2021. PMID: 33812315 Free PMC article.
-
Exploiting Hydrophobic Amino Acid Scanning to Develop Cyclic Peptide Inhibitors of the SARS-CoV-2 Main Protease with Antiviral Activity.Chemistry. 2024 Aug 6;30(44):e202401606. doi: 10.1002/chem.202401606. Epub 2024 Jul 17. Chemistry. 2024. PMID: 38801240
-
Computational guided identification of a citrus flavonoid as potential inhibitor of SARS-CoV-2 main protease.Mol Divers. 2021 Aug;25(3):1745-1759. doi: 10.1007/s11030-020-10150-x. Epub 2020 Nov 25. Mol Divers. 2021. PMID: 33236176 Free PMC article.
-
Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site.Viruses. 2020 Aug 26;12(9):942. doi: 10.3390/v12090942. Viruses. 2020. PMID: 32859008 Free PMC article.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
Cited by
-
Non-covalent cyclic peptides simultaneously targeting Mpro and NRP1 are highly effective against Omicron BA.2.75.Front Pharmacol. 2022 Nov 2;13:1037993. doi: 10.3389/fphar.2022.1037993. eCollection 2022. Front Pharmacol. 2022. PMID: 36408220 Free PMC article.
-
Large scale peptide screening against main protease of SARS CoV-2.J Comput Chem. 2023 Mar 30;44(8):887-901. doi: 10.1002/jcc.27050. Epub 2022 Dec 7. J Comput Chem. 2023. PMID: 36478400 Free PMC article.
-
In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2,4-amino acids targeting the SARS-CoV-2 main protease.Nat Chem. 2023 Jul;15(7):998-1005. doi: 10.1038/s41557-023-01205-1. Epub 2023 May 22. Nat Chem. 2023. PMID: 37217786 Free PMC article.
-
Structure-based drug design of an inhibitor of the SARS-CoV-2 (COVID-19) main protease using free software: A tutorial for students and scientists.Eur J Med Chem. 2021 Jun 5;218:113390. doi: 10.1016/j.ejmech.2021.113390. Epub 2021 Mar 20. Eur J Med Chem. 2021. PMID: 33812315 Free PMC article.
-
Antiviral Therapy of COVID-19.Int J Mol Sci. 2023 May 16;24(10):8867. doi: 10.3390/ijms24108867. Int J Mol Sci. 2023. PMID: 37240213 Free PMC article. Review.
References
-
- Sawyer T. CHAPTER 1:renaissance in peptide drug discovery: the third wave. in Peptide-based Drug Discovery: Challenges and New Therapeutics. 2017:1–34. doi: 10.1039/9781788011532-00001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

