Modulating FKBP5/FKBP51 and autophagy lowers HTT (huntingtin) levels
- PMID: 34024231
- PMCID: PMC8726715
- DOI: 10.1080/15548627.2021.1904489
Modulating FKBP5/FKBP51 and autophagy lowers HTT (huntingtin) levels
Abstract
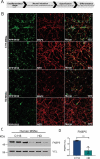
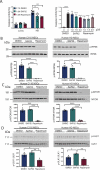
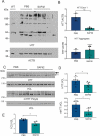
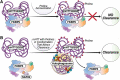
Current disease-modifying therapies for Huntington disease (HD) focus on lowering mutant HTT (huntingtin; mHTT) levels, and the immunosuppressant drug rapamycin is an intriguing therapeutic for aging and neurological disorders. Rapamycin interacts with FKBP1A/FKBP12 and FKBP5/FKBP51, inhibiting the MTORC1 complex and increasing cellular clearance mechanisms. Whether the levels of FKBP (FK506 binding protein) family members are altered in HD models and if these proteins are potential therapeutic targets for HD have not been investigated. Here, we found levels of FKBP5 are significantly reduced in HD R6/2 and zQ175 mouse models and human HD isogenic neural stem cells and medium spiny neurons derived from induced pluripotent stem cells. Moreover, FKBP5 interacts and colocalizes with HTT in the striatum and cortex of zQ175 mice and controls. Importantly, when we decreased FKBP5 levels or activity by genetic or pharmacological approaches, we observed reduced levels of mHTT in our isogenic human HD stem cell model. Decreasing FKBP5 levels by siRNA or pharmacological inhibition increased LC3-II levels and macroautophagic/autophagic flux, suggesting autophagic cellular clearance mechanisms are responsible for mHTT lowering. Unlike rapamycin, the effect of pharmacological inhibition with SAFit2, an inhibitor of FKBP5, is MTOR independent. Further, in vivo treatment for 2 weeks with SAFit2, results in reduced HTT levels in both HD R6/2 and zQ175 mouse models. Our studies establish FKBP5 as a protein involved in the pathogenesis of HD and identify FKBP5 as a potential therapeutic target for HD.Abbreviations : ACTB/β-actin: actin beta; AD: Alzheimer disease; BafA1: bafilomycin A1; BCA: bicinchoninic acid; BBB: blood brain barrier; BSA: bovine serum albumin; CoIP: co-immunoprecipitation; DMSO: dimethyl sulfoxide; DTT: dithiothreitol; FKBPs: FK506 binding proteins; HD: Huntington disease; HTT: huntingtin; iPSC: induced pluripotent stem cells; MAP1LC3/LC3:microtubule associated protein 1 light chain 3; MAPT/tau: microtubule associated protein tau; MES: 2-ethanesulfonic acid; MOPS: 3-(N-morphorlino)propanesulfonic acid); MSN: medium spiny neurons; mHTT: mutant huntingtin; MTOR: mechanistic target of rapamycin kinase; NSC: neural stem cells; ON: overnight; PD: Parkinson disease; PPIase: peptidyl-prolyl cis/trans-isomerases; polyQ: polyglutamine; PPP1R1B/DARPP-32: protein phosphatase 1 regulatory inhibitor subunit 1B; PTSD: post-traumatic stress disorder; RT: room temperature; SQSTM1/p62: sequestosome 1; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TBST:Tris-buffered saline, 0.1% Tween 20; TUBA: tubulin; ULK1: unc-51 like autophagy activating kinase 1; VCL: vinculin; WT: littermate controls.
Keywords: Autophagy; Huntington disease; SAFit2; fkbp12.6/fkbp1b; fkbp12/fkbp1a; fkbp51/fkbp5; fkbp52/fkbp4; induced pluripotent stem cells.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
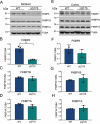

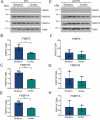
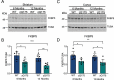

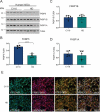


References
-
- Group THsDCR . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomesThe Huntington’s Disease Collaborative Research Group. Cell. 1993;72(6):971–983. - PubMed
-
- Hedreen JC, Peyser CE, Folstein SE, et al. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133(2):257–261. - PubMed
-
- Kiyama H, Seto-Ohshima A, Emson PC.. Calbindin D28K as a marker for the degeneration of the striatonigral pathway in Huntington’s disease. Brain Res. 1990;525(2):209–214. - PubMed
-
- Cardoso F. Differential diagnosis of Huntington’s disease: what the clinician should know. Neurodegener Dis Manag. 2014;4(1):67–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
