The Regulation of LexA on UV-Induced SOS Response in Myxococcus xanthus Based on Transcriptome Analysis
- PMID: 34024894
- PMCID: PMC9705874
- DOI: 10.4014/jmb.2103.03047
The Regulation of LexA on UV-Induced SOS Response in Myxococcus xanthus Based on Transcriptome Analysis
Abstract
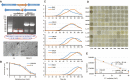
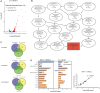
SOS response is a conserved response to DNA damage in prokaryotes and is negatively regulated by LexA protein, which recognizes specifically an "SOS-box" motif present in the promoter region of SOS genes. Myxococcus xanthus DK1622 possesses a lexA gene, and while the deletion of lexA had no significant effect on either bacterial morphology, UV-C resistance, or sporulation, it did delay growth. UV-C radiation resulted in 651 upregulated genes in M. xanthus, including the typical SOS genes lexA, recA, uvrA, recN and so on, mostly enriched in the pathways of DNA replication and repair, secondary metabolism, and signal transduction. The UV-irradiated lexA mutant also showed the induced expression of SOS genes and these SOS genes enriched into a similar pathway profile to that of wild-type strain. Without irradiation treatment, the absence of LexA enhanced the expression of 122 genes that were not enriched in any pathway. Further analysis of the promoter sequence revealed that in the 122 genes, only the promoters of recA2, lexA and an operon composed of three genes (pafB, pafC and cyaA) had SOS box sequence to which the LexA protein is bound directly. These results update our current understanding of SOS response in M. xanthus and show that UV induces more genes involved in secondary metabolism and signal transduction in addition to DNA replication and repair; and while the canonical LexA-dependent regulation on SOS response has shrunk, only 5 SOS genes are directly repressed by LexA.
Keywords: LexA; M. xanthus; SOS response; UV-C irradiation.
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

