Human Pathogenic Bacteria Detected in Rainwater: Risk Assessment and Correlation to Microbial Source Tracking Markers and Traditional Indicators
- PMID: 34025613
- PMCID: PMC8138566
- DOI: 10.3389/fmicb.2021.659784
Human Pathogenic Bacteria Detected in Rainwater: Risk Assessment and Correlation to Microbial Source Tracking Markers and Traditional Indicators
Abstract
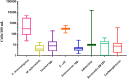
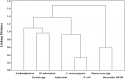
Roof-harvested rainwater (RHRW) was investigated for the presence of the human pathogenic bacteria Mycobacterium tuberculosis (M. tuberculosis), Yersinia spp. and Listeria monocytogenes (L. monocytogenes). While Yersinia spp. were detected in 92% (n = 25) of the RHRW samples, and L. monocytogenes and M. tuberculosis were detected in 100% (n = 25) of the samples, a significantly higher mean concentration (1.4 × 103 cells/100 mL) was recorded for L. monocytogenes over the sampling period. As the identification of appropriate water quality indicators is crucial to ensure access to safe water sources, correlation of the pathogens to traditional indicator organisms [Escherichia coli (E. coli) and Enterococcus spp.] and microbial source tracking (MST) markers (Bacteroides HF183, adenovirus and Lachnospiraceae) was conducted. A significant positive correlation was then recorded for E. coli versus L. monocytogenes (r = 0.6738; p = 0.000), and Enterococcus spp. versus the Bacteroides HF183 marker (r = 0.4071; p = 0.043), while a significant negative correlation was observed for M. tuberculosis versus the Bacteroides HF183 marker (r = -0.4558; p = 0.022). Quantitative microbial risk assessment indicated that the mean annual risk of infection posed by L. monocytogenes in the RHRW samples exceeded the annual infection risk benchmark limit (1 × 10-4 infections per person per year) for intentional drinking (∼10-4). In comparison, the mean annual risk of infection posed by E. coli was exceeded for intentional drinking (∼10-1), accidental consumption (∼10-3) and cleaning of the home (∼10-3). However, while the risk posed by M. tuberculosis for the two relevant exposure scenarios [garden hosing (∼10-5) and washing laundry by hand (∼10-5)] was below the benchmark limit, the risk posed by adenovirus for garden hosing (∼10-3) and washing laundry by hand (∼10-3) exceeded the benchmark limit. Thus, while the correlation analysis confirms that traditional indicators and MST markers should be used in combination to accurately monitor the pathogen-associated risk linked to the utilisation of RHRW, the integration of QMRA offers a more site-specific approach to monitor and estimate the human health risks associated with the use of RHRW.
Keywords: QMRA; human pathogenic bacteria; microbial source tracking markers; rainwater; traditional indicator organisms.
Copyright © 2021 Denissen, Reyneke, Waso, Khan and Khan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Microbial source tracking markers associated with domestic rainwater harvesting systems: Correlation to indicator organisms.Environ Res. 2018 Feb;161:446-455. doi: 10.1016/j.envres.2017.11.043. Environ Res. 2018. PMID: 29216491
-
Human health risks for Legionella and Mycobacterium avium complex (MAC) from potable and non-potable uses of roof-harvested rainwater.Water Res. 2017 Aug 1;119:288-303. doi: 10.1016/j.watres.2017.04.004. Epub 2017 Apr 5. Water Res. 2017. PMID: 28500949
-
Abundance of Naegleria fowleri in roof-harvested rainwater tank samples from two continents.Environ Sci Pollut Res Int. 2018 Feb;25(6):5700-5710. doi: 10.1007/s11356-017-0870-9. Epub 2017 Dec 11. Environ Sci Pollut Res Int. 2018. PMID: 29230646
-
Microbiological quality of roof-harvested rainwater and health risks: a review.J Environ Qual. 2011 Jan-Feb;40(1):13-21. doi: 10.2134/jeq2010.0345. J Environ Qual. 2011. PMID: 21488488 Review.
-
Synergy between quantitative microbial source tracking (qMST) and quantitative microbial risk assessment (QMRA): A review and prospectus.Environ Int. 2019 Sep;130:104703. doi: 10.1016/j.envint.2019.03.051. Epub 2019 Jul 8. Environ Int. 2019. PMID: 31295713 Review.
Cited by
-
Microbial Characterization, Factors Contributing to Contamination, and Household Use of Cistern Water, U.S. Virgin Islands.ACS ES T Water. 2022 Dec 9;2(12):2634-2644. doi: 10.1021/acsestwater.2c00389. Epub 2022 Oct 18. ACS ES T Water. 2022. PMID: 36530952 Free PMC article.
-
Variations in Bacterial Communities and Antibiotic Resistance Genes Across Diverse Recycled and Surface Water Irrigation Sources in the Mid-Atlantic and Southwest United States: A CONSERVE Two-Year Field Study.Environ Sci Technol. 2022 Nov 1;56(21):15019-15033. doi: 10.1021/acs.est.2c02281. Epub 2022 Oct 4. Environ Sci Technol. 2022. PMID: 36194536 Free PMC article.
-
Quantitative microbial risk assessment of pathogen exposure from rainwater used in high-pressure vehicle washing.J Water Health. 2025 Mar;23(3):428-438. doi: 10.2166/wh.2025.365. Epub 2025 Mar 1. J Water Health. 2025. PMID: 40156219
References
-
- Ahmed W., Goonetilleke A., Gardner T. (2008a). Alternative indicators for detection and quantification of faecal pollution. Water (Australia) 39 46–49. 10.HH/j.l472-765X.2007.02287.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

