Contribution of Different Mechanisms to Ciprofloxacin Resistance in Salmonella spp
- PMID: 34025618
- PMCID: PMC8137344
- DOI: 10.3389/fmicb.2021.663731
Contribution of Different Mechanisms to Ciprofloxacin Resistance in Salmonella spp
Abstract
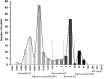
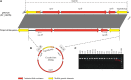
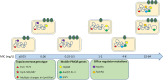
Development of fluoroquinolone resistance can involve several mechanisms that include chromosomal mutations in genes (gyrAB and parCE) encoding the target bacterial topoisomerase enzymes, increased expression of the AcrAB-TolC efflux system, and acquisition of transmissible quinolone-resistance genes. In this study, 176 Salmonella isolates from animals with a broad range of ciprofloxacin MICs were collected to analyze the contribution of these different mechanisms to different phenotypes. All isolates were classified according to their ciprofloxacin susceptibility pattern into five groups as follows: highly resistant (HR), resistant (R), intermediate (I), reduced susceptibility (RS), and susceptible (S). We found that the ParC T57S substitution was common in strains exhibiting lowest MICs of ciprofloxacin while increased MICs depended on the type of GyrA mutation. The ParC T57S substitution appeared to incur little cost to bacterial fitness on its own. The presence of PMQR genes represented an route for resistance development in the absence of target-site mutations. Switching of the plasmid-mediated quinolone resistance (PMQR) gene location from a plasmid to the chromosome was observed and resulted in decreased ciprofloxacin susceptibility; this also correlated with increased fitness and a stable resistance phenotype. The overexpression of AcrAB-TolC played an important role in isolates with small decreases in susceptibility and expression was upregulated by MarA more often than by RamA. This study increases our understanding of the relative importance of several resistance mechanisms in the development of fluoroquinolone resistance in Salmonella from the food chain.
Keywords: AcrAB efflux pump; PMQR; QRDR; Salmonella; circular intermediate; fluoroquinolone resistance.
Copyright © 2021 Chang, Zhang, Sun, Li, Lin, Yang, Webber and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Analysis of the Fluoroquinolone Antibiotic Resistance Mechanism of Salmonella enterica Isolates.J Microbiol Biotechnol. 2016 Sep 28;26(9):1605-12. doi: 10.4014/jmb.1602.02063. J Microbiol Biotechnol. 2016. PMID: 27116992
-
'Supermutators' found amongst highly levofloxacin-resistant E. coli isolates: a rapid protocol for the detection of mutation sites.Emerg Microbes Infect. 2015 Jan;4(1):e4. doi: 10.1038/emi.2015.4. Epub 2015 Jan 28. Emerg Microbes Infect. 2015. PMID: 26038761 Free PMC article.
-
Molecular and Physiological Characterization of Fluoroquinolone-Highly Resistant Salmonella Enteritidis Strains.Front Microbiol. 2019 Apr 9;10:729. doi: 10.3389/fmicb.2019.00729. eCollection 2019. Front Microbiol. 2019. PMID: 31024504 Free PMC article.
-
Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium.J Antimicrob Chemother. 2009 Jan;63(1):95-102. doi: 10.1093/jac/dkn448. Epub 2008 Nov 4. J Antimicrob Chemother. 2009. PMID: 18984645
-
Mechanisms of fluoroquinolone resistance: an update 1994-1998.Drugs. 1999;58 Suppl 2:11-8. doi: 10.2165/00003495-199958002-00003. Drugs. 1999. PMID: 10553699 Review.
Cited by
-
Whole-genome sequencing-based characterization of Salmonella enterica Serovar Enteritidis and Kentucky isolated from laying hens in northwest of Iran, 2022-2023.Gut Pathog. 2025 Jan 16;17(1):2. doi: 10.1186/s13099-025-00679-3. Gut Pathog. 2025. PMID: 39819347 Free PMC article.
-
Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan.Antibiotics (Basel). 2021 Nov 11;10(11):1383. doi: 10.3390/antibiotics10111383. Antibiotics (Basel). 2021. PMID: 34827321 Free PMC article.
-
Clonal Spread and Genetic Mechanisms Underpinning Ciprofloxacin Resistance in Salmonella enteritidis.Foods. 2025 Jan 16;14(2):289. doi: 10.3390/foods14020289. Foods. 2025. PMID: 39856955 Free PMC article.
-
Genomic and Transcriptomic Analysis of Bovine Pasteurella multocida Serogroup A Strain Reveals Insights Into Virulence Attenuation.Front Vet Sci. 2021 Nov 10;8:765495. doi: 10.3389/fvets.2021.765495. eCollection 2021. Front Vet Sci. 2021. PMID: 34859092 Free PMC article.
-
Prevalence, antimicrobial resistance, and genomic characterization of Salmonella strains isolated in Hangzhou, China: a two-year study.Ann Clin Microbiol Antimicrob. 2024 Sep 28;23(1):86. doi: 10.1186/s12941-024-00748-6. Ann Clin Microbiol Antimicrob. 2024. PMID: 39342293 Free PMC article.
References
-
- Barry K. E., Wailan A. M., Sheppard A. E., Crook D., Vegesana K., Stoesser N., et al. (2019). Don’t overlook the little guy: an evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid 103 1–8. 10.1016/j.plasmid.2019.03.005 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

