Introduced populations of ragweed show as much evolutionary potential as native populations
- PMID: 34025777
- PMCID: PMC8127702
- DOI: 10.1111/eva.13211
Introduced populations of ragweed show as much evolutionary potential as native populations
Abstract
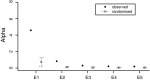
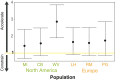
Invasive species are a global economic and ecological problem. They also offer an opportunity to understand evolutionary processes in a colonizing context. The impacts of evolutionary factors, such as genetic variation, on the invasion process are increasingly appreciated, but there remain gaps in the empirical literature. The adaptive potential of populations can be quantified using genetic variance-covariance matrices (G), which encapsulate the heritable genetic variance in a population. Here, we use a multivariate Bayesian approach to assess the adaptive potential of invasive populations of ragweed (Ambrosia artemisiifolia), a serious allergen and agricultural weed. We compared several aspects of genetic architecture and the structure of G matrices between three native and three introduced populations, based on phenotypic data collected in a field common garden experiment. We found moderate differences in the quantitative genetic architecture among populations, but we did not find that introduced populations suffer from a limited adaptive potential or increased genetic constraint compared with native populations. Ragweed has an annual life history, is an obligate outcrosser, and produces very large numbers of seeds and pollen grains. These characteristics, combined with the significant additive genetic variance documented here, suggest ragweed will be able to respond quickly to selection pressures in both its native and introduced ranges.
Keywords: Ambrosia artemisiifolia; G matrices; additive genetic variance; introduced species.
© 2021 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bacigalupe, L. D. (2009). Biological invasions and phenotypic evolution: A quantitative genetic perspective. Biological Invasions, 11, 2243–2250.
-
- Barrett, S. C. H. (2015). Foundations of invasion genetics: The Baker and Stebbins legacy. Molecular Ecology, 24, 1927–1941. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

