Silencing LINC01021 inhibits gastric cancer through upregulation of KISS1 expression by blocking CDK2-dependent phosphorylation of CDX2
- PMID: 34026327
- PMCID: PMC8121629
- DOI: 10.1016/j.omtn.2021.01.025
Silencing LINC01021 inhibits gastric cancer through upregulation of KISS1 expression by blocking CDK2-dependent phosphorylation of CDX2
Abstract
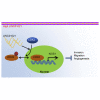
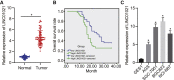
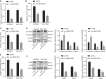
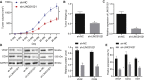
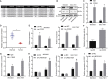
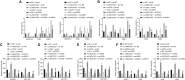
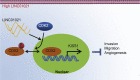
Gastric cancer remains one of the most dangerous cancers, bringing suffering and economic burden to people worldwide. Long noncoding RNAs (lncRNAs) exhibit great potentials for targeted therapy of various cancers. In this investigation, we tested mechanisms by which LINC01021 may regulate gastric cancer progression. We collected gastric cancer tissues and procured cell lines to explore the potential factors by which LINC01021 had effects on angiogenesis, invasion, and migration, by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), Transwell assay, and western blot analysis. Relationships among LINC01021, Caudal-type homeobox 2 (CDX2), and KISS1 were validated by dual-luciferase gene reporter, RNA pull-down, and RNA immunoprecipitation assays. Additionally, a murine model was developed to further explore the impact of LINC01021 on tumors in vivo. LINC01021 was upregulated in gastric cancer tissues and cells. LINC01021 regulated KISS1 through CDK2, which promoted phosphorylation and nuclear export in CDX2. Inhibition of LINC01021 suppressed the tumorigenesis of gastric cancer. Further, silencing LINC01021 exerted an inhibitory effect on cancer cell migration, invasion, and angiogenesis by promoting the binding between CDX2 and KISS1, while inhibiting that between CDK2 and CDX2. Taken altogether, high LINC01021 expression in gastric cancer promotes malignant cell migration and angiogenesis by downregulation of KISS1 through CDK2-mediated CDX2 phosphorylation.
Keywords: CDK2; CDX2; KISS1; LINC01021; gastric cancer; phosphorylation.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Thrumurthy S.G., Chaudry M.A., Chau I., Allum W. Does surgery have a role in managing incurable gastric cancer? Nat. Rev. Clin. Oncol. 2015;12:676–682. - PubMed
-
- Carlevaro-Fita J., Johnson R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell. 2019;73:869–883. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

