A Fifteen-Gene Classifier to Predict Neoadjuvant Chemotherapy Responses in Patients with Stage IB to IIB Squamous Cervical Cancer
- PMID: 34026427
- PMCID: PMC8132153
- DOI: 10.1002/advs.202001978
A Fifteen-Gene Classifier to Predict Neoadjuvant Chemotherapy Responses in Patients with Stage IB to IIB Squamous Cervical Cancer
Abstract
Neoadjuvant chemotherapy (NACT) remains an attractive alternative for controlling locally advanced cervical cancer. However, approximately 15-34% of women do not respond to induction therapy. To develop a risk stratification tool, 56 patients with stage IB-IIB cervical cancer are included in 2 research centers from the discovery cohort. Patient-specific somatic mutations led to NACT non-responsiveness are identified by whole-exome sequencing. Next, CRISPR/Cas9-based library screenings are performed based on these genes to confirm their biological contribution to drug resistance. A 15-gene classifier is developed by generalized linear regression analysis combined with the logistic regression model. In an independent validation cohort of 102 patients, the classifier showed good predictive ability with an area under the curve of 0.80 (95% confidence interval (CI), 0.69-0.91). Furthermore, the 15-gene classifier is significantly associated with patient responsiveness to NACT in both univariate (odds ratio, 10.8; 95% CI, 3.55-32.86; p = 2.8 × 10-5) and multivariate analysis (odds ratio, 17.34; 95% CI, 4.04-74.40; p = 1.23 × 10-4) in the validation set. In conclusion, the 15-gene classifier can accurately predict the clinical response to NACT before treatment, representing a promising approach for guiding the selection of appropriate treatment strategies for locally advanced cervical cancer.
Keywords: CRISPR/Cas9‐based library screening; neoadjuvant chemotherapy; precision medicine; whole exon sequencing.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

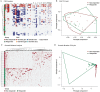

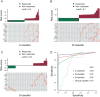


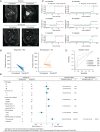
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Ca‐Cancer J. Clin. 2018, 68, 394. - PubMed
-
- Abu‐Rustum N. R., Yashar C. M., Bean S., Bradley K., Campos S. M., Chon H. S., Chu C., Cohn D., Crispens M. A., Damast S., Fisher C. M., Frederick P., Gaffney D. K., Giuntoli R., Han E., Huh W. K., Lurain J. R. III, Mariani A., Mutch D., Nagel C., Nekhlyudov L., Fader A. N., Remmenga S. W., Reynolds R. K., Sisodia R., Tillmanns T., Ueda S., Urban R., Wyse E., McMillian N. R., Motter A. D., J. Natl. Compr. Canc. Netw. 2020, 18, 660. - PubMed
-
- Wo J. Y., Viswanathan A. N., Int. J. Radiat. Oncol., Biol., Phys. 2009, 73, 1304.
-
- Gaffney D. K., Du Bois A., Narayan K., Reed N., Toita T., Pignata S., Blake P., Portelance L., Sadoyze A., Potter R., Colombo A., Randall M., Mirza M. R., Trimble E. L., Int. J. Radiat. Oncol., Biol., Phys. 2007, 68, 485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
