Dual Inhibition of DKC1 and MEK1/2 Synergistically Restrains the Growth of Colorectal Cancer Cells
- PMID: 34026451
- PMCID: PMC8132060
- DOI: 10.1002/advs.202004344
Dual Inhibition of DKC1 and MEK1/2 Synergistically Restrains the Growth of Colorectal Cancer Cells
Abstract
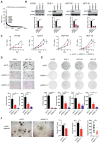
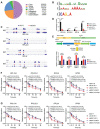
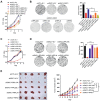
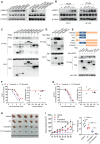
Colorectal cancer, one of the most commonly diagnosed cancers worldwide, is often accompanied by uncontrolled proliferation of tumor cells. Dyskerin pseudouridine synthase 1 (DKC1), screened using the genome-wide RNAi strategy, is a previously unidentified key regulator that promotes colorectal cancer cell proliferation. Enforced expression of DKC1, but not its catalytically inactive mutant D125A, accelerates cell growth in vitro and in vivo. DKC1 knockdown or its inhibitor pyrazofurin attenuates cell proliferation. Proteomics, RNA immunoprecipitation (RIP)-seq, and RNA decay analyses reveal that DKC1 binds to and stabilizes the mRNA of several ribosomal proteins (RPs), including RPL10A, RPL22L1, RPL34, and RPS3. DKC1 depletion significantly accelerates mRNA decay of these RPs, which mediates the oncogenic function of DKC1. Interestingly, these DKC1-regulated RPs also interact with HRAS and suppress the RAS/RAF/MEK/ERK pathway. Pyrazofurin and trametinib combination synergistically restrains colorectal cancer cell growth in vitro and in vivo. Furthermore, DKC1 is markedly upregulated in colorectal cancer tissues compared to adjacent normal tissues. Colorectal cancer patients with higher DKC1 expression has consistently poorer overall survival and progression-free survival outcomes. Taken together, these data suggest that DKC1 is an essential gene and candidate therapeutic target for colorectal cancer.
Keywords: MEK1/2; dyskerin pseudouridine synthase 1; pyrazofurin; ribosomal protein; trametinib.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Siegel R. L., Miller K. D., Jemal A., Ca‐Cancer J. Clin. 2020, 70, 7. - PubMed
-
- Cremolini C., Antoniotti C., Rossini D., Lonardi S., Loupakis F., Pietrantonio F., Bordonaro R., Latiano T. P., Tamburini E., Santini D., Passardi A., Marmorino F., Grande R., Aprile G., Zaniboni A., Murgioni S., Granetto C., Buonadonna A., Moretto R., Corallo S., Cordio S., Antonuzzo L., Tomasello G., Masi G., Ronzoni M., Di Donato S., Carlomagno C., Clavarezza M., Ritorto G., Mambrini A., Roselli M., Cupini S., Mammoliti S., Fenocchio E., Corgna E., Zagonel V., Fontanini G., Ugolini C., Boni L., Falcone A., Lancet Oncol. 2020, 21, 497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
