Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: Molecular docking, molecular dynamics, and SAR studies
- PMID: 34026584
- PMCID: PMC8126476
- DOI: 10.1016/j.jtcme.2021.05.001
Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: Molecular docking, molecular dynamics, and SAR studies
Abstract
Background and aim: The discovery of drugs capable of inhibiting SARS-CoV-2 is a priority for human beings due to the severity of the global health pandemic caused by COVID-19. To this end, natural products can provide therapeutic alternatives that could be employed as an effective safe treatment for COVID-19.
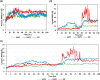
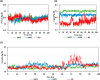
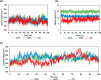
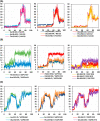
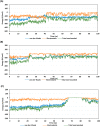
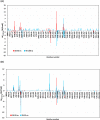
Experimental procedure: Twelve compounds were isolated from the aerial parts of C. officinalis L. and investigated for their inhibitory activities against SARS-CoV-2 Mpro compared to its co-crystallized N3 inhibitor using molecular docking studies. Furthermore, a 100 ns MD simulation was performed for the most active two promising compounds, Calendulaglycoside A (SAP5) and Osteosaponin-I (SAP8).
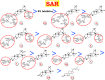
Results and conclusion: At first, molecular docking studies showed interesting binding scores as compared to the N3 inhibitor. Calendulaglycoside A (SAP5) achieved a superior binding than the co-crystallized inhibitor indicating promising affinity and intrinsic activity towards the Mpro of SARS-CoV-2 as well. Moreover, findings illustrated preferential stability for SAP5 within the Mpro pocket over that of N3 beyond the 40 ns MD simulation course. Structural preferentiality for triterpene-Mpro binding highlights the significant role of 17β-glucosyl and carboxylic 3α-galactosyl I moieties through high electrostatic interactions across the MD simulation trajectories. Furthermore, this study clarified a promising SAR responsible for the antiviral activity against the SARS-CoV-2 Mpro and the design of new drug candidates targeting it as well. The above findings could be promising for fast examining the previously isolated triterpenes both pre-clinically and clinically for the treatment of COVID-19.
Keywords: C. officinalis L.; COVID-19; Computational studies; SAR; Triterpenes.
© 2021 Center for Food and Biomolecules, National Taiwan University. Production and hosting by Elsevier Taiwan LLC.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
