YTHDF1 Is a Potential Pan-Cancer Biomarker for Prognosis and Immunotherapy
- PMID: 34026603
- PMCID: PMC8134747
- DOI: 10.3389/fonc.2021.607224
YTHDF1 Is a Potential Pan-Cancer Biomarker for Prognosis and Immunotherapy
Abstract
Background: YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) has been indicated proven to participate in the cross-presentation of tumor antigens in dendritic cells and the cross-priming of CD8+ T cells. However, the role of YTHDF1 in prognosis and immunology in human cancers remains largely unknown.
Methods: All original data were downloaded from TCGA and GEO databases and integrated via R 3.2.2. YTHDF1 expression was explored with the Oncomine, TIMER, GEPIA, and BioGPS databases. The effect of YTHDF1 on prognosis was analyzed via GEPIA, Kaplan-Meier plotter, and the PrognoScan database. The TISIDB database was used to determine YTHDF1 expression in different immune and molecular subtypes of human cancers. The correlations between YTHDF1 expression and immune checkpoints (ICP), tumor mutational burden (TMB), microsatellite instability (MSI), and neoantigens in human cancers were analyzed via the SangerBox database. The relationships between YTHDF1 expression and tumor-infiltrated immune cells were analyzed via the TIMER and GEPIA databases. The relationships between YTHDF1 and marker genes of tumor-infiltrated immune cells in urogenital cancers were analyzed for confirmation. The genomic alterations of YTHDF1 were investigated with the c-BioPortal database. The differential expression of YTHDF1 in urogenital cancers with different clinical characteristics was analyzed with the UALCAN database. YTHDF1 coexpression networks were studied by the LinkedOmics database.
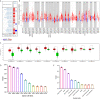
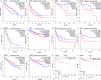
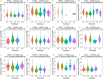
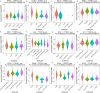
Results: In general, YTHDF1 expression was higher in tumors than in paired normal tissue in human cancers. YTHDF1 expression had strong relationships with prognosis, ICP, TMB, MSI, and neoantigens. YTHDF1 plays an essential role in the tumor microenvironment (TME) and participates in immune regulation. Furthermore, significant strong correlations between YTHDF1 expression and tumor immune-infiltrated cells (TILs) existed in human cancers, and marker genes of TILs were significantly related to YTHDF expression in urogenital cancers. TYHDF1 coexpression networks mostly participated in the regulation of immune response and antigen processing and presentation.
Conclusion: YTHDF1 may serve as a potential prognostic and immunological pan-cancer biomarker. Moreover, YTHDF1 could be a novel target for tumor immunotherapy.
Keywords: YTHDF1; human cancer; immune infiltration; immunotherapy; prognosis; tumor microenvironment.
Copyright © 2021 Hu, Qiu, Yu, Hu, Deng, Li, Yi, Chen and Zu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

