Alterations of Gut Microbiota in Patients With Graves' Disease
- PMID: 34026662
- PMCID: PMC8132172
- DOI: 10.3389/fcimb.2021.663131
Alterations of Gut Microbiota in Patients With Graves' Disease
Abstract
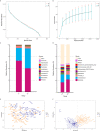
Graves' disease (GD) is a systemic autoimmune disease characterized by hyperthyroidism. Evidence suggests that alterations to the gut microbiota may be involved in the development of autoimmune disorders. The aim of this study was to characterize the composition of gut microbiota in GD patients. Fecal samples were collected from 55 GD patients and 48 healthy controls. Using 16S rRNA gene amplification and sequencing, the overall bacterial richness and diversity were found to be similar between GD patients and healthy controls. However, principal coordinate analysis and partial least squares-discriminant analysis showed that the overall gut microbiota composition was significantly different (ANOSIM; p < 0.001). The linear discriminant analysis effect size revealed that Firmicutes phylum decreased in GD patients, with a corresponding increase in Bacteroidetes phylum compared to healthy controls. In addition, the families Prevotellaceae, and Veillonellaceae and the genus Prevotella_9 were closely associated with GD patients, while the families Lachnospiraceae and Ruminococcaceae and the genera Faecalibacterium, Lachnospira, and Lachnospiraceae NK4A136 were associated with healthy controls. Metagenomic profiles analysis yielded 22 statistically significant bacterial taxa: 18 taxa were increased and 4 taxa were decreased. Key bacterial taxa with different abundances between the two groups were strongly correlated with GD-associated clinical parameters using Spearman's correlation analysis. Importantly, the discriminant model based on predominant microbiota could effectively distinguish GD patients from healthy controls (AUC = 0.825). Thus, the gut microbiota composition between GD patients and healthy controls is significantly difference, indicating that gut microbiota may play a role in the pathogenesis of GD. Further studies are needed to fully elucidate the role of gut microbiota in the development of GD.
Keywords: 16S rRNA; Graves’ disease; clinical parameters; gut microbiota; next-generation sequencing.
Copyright © 2021 Chang, Lin, Chen, Chang, Yeh, Lo and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

