ML-AdVInfect: A Machine-Learning Based Adenoviral Infection Predictor
- PMID: 34026828
- PMCID: PMC8139618
- DOI: 10.3389/fmolb.2021.647424
ML-AdVInfect: A Machine-Learning Based Adenoviral Infection Predictor
Abstract
Adenoviruses (AdVs) constitute a diverse family with many pathogenic types that infect a broad range of hosts. Understanding the pathogenesis of adenoviral infections is not only clinically relevant but also important to elucidate the potential use of AdVs as vectors in therapeutic applications. For an adenoviral infection to occur, attachment of the viral ligand to a cellular receptor on the host organism is a prerequisite and, in this sense, it is a criterion to decide whether an adenoviral infection can potentially happen. The interaction between any virus and its corresponding host organism is a specific kind of protein-protein interaction (PPI) and several experimental techniques, including high-throughput methods are being used in exploring such interactions. As a result, there has been accumulating data on virus-host interactions including a significant portion reported at publicly available bioinformatics resources. There is not, however, a computational model to integrate and interpret the existing data to draw out concise decisions, such as whether an infection happens or not. In this study, accepting the cellular entry of AdV as a decisive parameter for infectivity, we have developed a machine learning, more precisely support vector machine (SVM), based methodology to predict whether adenoviral infection can take place in a given host. For this purpose, we used the sequence data of the known receptors of AdVs, we identified sets of adenoviral ligands and their respective host species, and eventually, we have constructed a comprehensive adenovirus-host interaction dataset. Then, we committed interaction predictions through publicly available virus-host PPI tools and constructed an AdV infection predictor model using SVM with RBF kernel, with the overall sensitivity, specificity, and AUC of 0.88 ± 0.011, 0.83 ± 0.064, and 0.86 ± 0.030, respectively. ML-AdVInfect is the first of its kind as an effective predictor to screen the infection capacity along with anticipating any cross-species shifts. We anticipate our approach led to ML-AdVInfect can be adapted in making predictions for other viral infections.
Keywords: PPI prediction; adenovirus; host susceptibility; host-pathogen interaction; viral infection prediction; virus bioinformatics; virus-host interaction.
Copyright © 2021 Karabulut, Karpuzcu, Türk, Ibrahim and Süzek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.This project is supported by The Scientific and Technological Research Council of Turkey (Grant number: 119E664) We would like to acknowledge: Türkiye Bilimsel ve Teknolojik Araştirma Kurumu (Award number(s): 119E664).
Figures
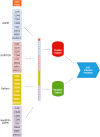
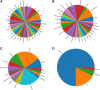
References
-
- Author Anonymous (2020a). HAdV Working Group [Online]. Available: http://hadvwg.gmu.edu (Accessed December 24, 2020)
-
- Author Anonymous (2020b). Scikit-Learn: Machine Learning in Python — Scikit-Learn 0.24.0 Documentation [Online]. Available: https://scikit-learn.org/stable (Accessed December 24, 2020)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

