Avadomide induces degradation of ZMYM2 fusion oncoproteins in hematologic malignancies
- PMID: 34027417
- PMCID: PMC8133393
- DOI: 10.1158/2643-3230.BCD-20-0105
Avadomide induces degradation of ZMYM2 fusion oncoproteins in hematologic malignancies
Abstract
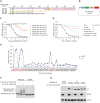
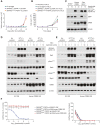
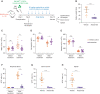
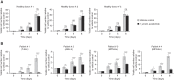
Thalidomide analogs exert their therapeutic effects by binding to the CRL4CRBN E3 ubiquitin ligase, promoting ubiquitination and subsequent proteasomal degradation of specific protein substrates. Drug-induced degradation of IKZF1 and IKZF3 in B-cell malignancies demonstrates the clinical utility of targeting disease-relevant transcription factors for degradation. Here, we found that avadomide (CC-122) induces CRBN-dependent ubiquitination and proteasomal degradation of ZMYM2 (ZNF198), a transcription factor involved in balanced chromosomal rearrangements with FGFR1 and FLT3 in aggressive forms of hematologic malignancies. The minimal drug-responsive element of ZMYM2 is a zinc-chelating MYM domain and is contained in the N-terminal portion of ZMYM2 that is universally included in the derived fusion proteins. We demonstrate that avadomide has the ability to induce proteasomal degradation of ZMYM2-FGFR1 and ZMYM2-FLT3 chimeric oncoproteins, both in vitro and in vivo. Our findings suggest that patients with hematologic malignancies harboring these ZMYM2 fusion proteins may benefit from avadomide treatment.
Keywords: fusion oncoproteins; hematologic malignancies; thalidomide analogs; ubiquitination; zinc finger protein.
Conflict of interest statement
Disclosure of potential conflicts of interest B.L. Ebert has received research funding from Celgene and Deerfield. He has received consulting fees from GRAIL, and he serves on the scientific advisory boards for Skyhawk Therapeutics, Exo Therapeutics, and Neomorph Therapeutics. S.A. Carr is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and a scientific advisor to Pfizer and Biogen.
Figures



Similar articles
-
Targeting cereblon in hematologic malignancies.Blood Rev. 2023 Jan;57:100994. doi: 10.1016/j.blre.2022.100994. Epub 2022 Jul 31. Blood Rev. 2023. PMID: 35933246 Review.
-
Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.Science. 2018 Nov 2;362(6414):eaat0572. doi: 10.1126/science.aat0572. Science. 2018. PMID: 30385546 Free PMC article.
-
Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase.Nature. 2016 Apr 7;532(7597):127-30. doi: 10.1038/nature16979. Epub 2016 Feb 24. Nature. 2016. PMID: 26909574
-
Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide.Nature. 2014 Aug 7;512(7512):49-53. doi: 10.1038/nature13527. Epub 2014 Jul 16. Nature. 2014. PMID: 25043012 Free PMC article.
-
The molecular mechanism of thalidomide analogs in hematologic malignancies.J Mol Med (Berl). 2016 Dec;94(12):1327-1334. doi: 10.1007/s00109-016-1450-z. Epub 2016 Aug 5. J Mol Med (Berl). 2016. PMID: 27492707 Review.
Cited by
-
Hyperthermia promotes degradation of the acute promyelocytic leukemia driver oncoprotein ZBTB16/RARα.Acta Pharmacol Sin. 2023 Apr;44(4):822-831. doi: 10.1038/s41401-022-01001-6. Epub 2022 Oct 10. Acta Pharmacol Sin. 2023. PMID: 36216898 Free PMC article.
-
Targeted Protein Degradation: An Important Tool for Drug Discovery for "Undruggable" Tumor Transcription Factors.Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221095950. doi: 10.1177/15330338221095950. Technol Cancer Res Treat. 2022. PMID: 35466792 Free PMC article.
-
Molecular glue degrader for tumor treatment.Front Oncol. 2024 Dec 12;14:1512666. doi: 10.3389/fonc.2024.1512666. eCollection 2024. Front Oncol. 2024. PMID: 39759140 Free PMC article. Review.
-
Novel potent molecular glue degraders against broad range of hematological cancer cell lines via multiple neosubstrates degradation.J Hematol Oncol. 2024 Sep 2;17(1):77. doi: 10.1186/s13045-024-01592-z. J Hematol Oncol. 2024. PMID: 39218923 Free PMC article.
-
Targeting the ZMYM2-ANXA9 Axis with FLT3 Inhibitor G749 Overcomes Oxaliplatin Resistance in Colorectal Cancer.Biomedicines. 2025 May 20;13(5):1247. doi: 10.3390/biomedicines13051247. Biomedicines. 2025. PMID: 40427072 Free PMC article.
References
-
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, et al. Identification of a primary target of thalidomide teratogenicity. Science 2010;327:1345–50. - PubMed
-
- Chamberlain PP, Cathers BE. Cereblon modulators: low molecular weight inducers of protein degradation. Drug Discov Today Technol 2019;31:29–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

