Highly specific monoclonal antibodies and epitope identification against SARS-CoV-2 nucleocapsid protein for antigen detection tests
- PMID: 34027498
- PMCID: PMC8126173
- DOI: 10.1016/j.xcrm.2021.100311
Highly specific monoclonal antibodies and epitope identification against SARS-CoV-2 nucleocapsid protein for antigen detection tests
Abstract
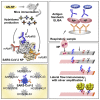
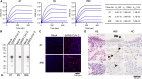
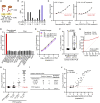
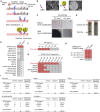
The ongoing coronavirus disease 2019 (COVID-19) pandemic is a major global public health concern. Although rapid point-of-care testing for detecting viral antigen is important for management of the outbreak, the current antigen tests are less sensitive than nucleic acid testing. In our current study, we produce monoclonal antibodies (mAbs) that exclusively react with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and exhibit no cross-reactivity with other human coronaviruses, including SARS-CoV. Molecular modeling suggests that the mAbs bind to epitopes present on the exterior surface of the nucleocapsid, making them suitable for detecting SARS-CoV-2 in clinical samples. We further select the optimal pair of anti-SARS-CoV-2 nucleocapsid protein (NP) mAbs using ELISA and then use this mAb pair to develop immunochromatographic assay augmented with silver amplification technology. Our mAbs recognize the variants of concern (501Y.V1-V3) that are currently in circulation. Because of their high performance, the mAbs of this study can serve as good candidates for developing antigen detection kits for COVID-19.
Keywords: COVID-19; SARS-CoV-2; monoclonal antibody; nucleoprotein; point-of-care testing.
© 2021 The Authors.
Conflict of interest statement
Y. Yamaoka, S.K., K. Suzuki, and D.A. are current employees of Kanto Chemical Co., Inc.; J.K., A.W., and T.T. are current employees of FUJIFILM Corporation. A.R. received collaborative research grant from Kanto Chemical Co., Inc. and FUJIFILM Corporation. Provisional patent applications relevant to this study were filed. The remaining authors declare no competing interests.
Figures

Similar articles
-
Preparation of highly specific monoclonal antibodies against SARS-CoV-2 nucleocapsid protein and the preliminary development of antigen detection test strips.J Med Virol. 2022 Apr;94(4):1633-1640. doi: 10.1002/jmv.27520. Epub 2021 Dec 21. J Med Virol. 2022. PMID: 34904253 Free PMC article.
-
Monoclonal Antibodies against Nucleocapsid Protein of SARS-CoV-2 Variants for Detection of COVID-19.Int J Mol Sci. 2021 Nov 17;22(22):12412. doi: 10.3390/ijms222212412. Int J Mol Sci. 2021. PMID: 34830291 Free PMC article.
-
Immunoassay Detection of SARS-CoV-2 Using Monoclonal Antibody Binding to Viral Nucleocapsid Protein.Microb Biotechnol. 2025 Feb;18(2):e70117. doi: 10.1111/1751-7915.70117. Microb Biotechnol. 2025. PMID: 39989430 Free PMC article.
-
Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective.J Clin Microbiol. 2021 Apr 20;59(5):e02160-20. doi: 10.1128/JCM.02160-20. Print 2021 Apr 20. J Clin Microbiol. 2021. PMID: 33318065 Free PMC article. Review.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
Cited by
-
Development of monoclonal antibodies against SARS-CoV-2 nucleocapsid protein for COVID-19 antigen detection.Trop Med Health. 2025 May 13;53(1):69. doi: 10.1186/s41182-025-00756-y. Trop Med Health. 2025. PMID: 40361217 Free PMC article.
-
Rapid Biosensor of SARS-CoV-2 Using Specific Monoclonal Antibodies Recognizing Conserved Nucleocapsid Protein Epitopes.Viruses. 2022 Jan 27;14(2):255. doi: 10.3390/v14020255. Viruses. 2022. PMID: 35215848 Free PMC article.
-
Optical lateral flow assays in early diagnosis of SARS-CoV-2 infection.Anal Sci. 2024 Sep;40(9):1571-1591. doi: 10.1007/s44211-024-00596-6. Epub 2024 May 17. Anal Sci. 2024. PMID: 38758251 Review.
-
Characterization and Utilization of Disulfide-Bonded SARS-CoV-2 Receptor Binding Domain of Spike Protein Synthesized by Wheat Germ Cell-Free Production System.Viruses. 2022 Jul 1;14(7):1461. doi: 10.3390/v14071461. Viruses. 2022. PMID: 35891441 Free PMC article.
-
Patient-derived monoclonal antibodies to SARS-CoV-2 nucleocapsid protein N-terminal and C-terminal domains cross-react with their counterparts of SARS-CoV, but not other human betacoronaviruses.Front Immunol. 2023 Jan 31;14:1093709. doi: 10.3389/fimmu.2023.1093709. eCollection 2023. Front Immunol. 2023. PMID: 36798118 Free PMC article.
References
-
- World Health Organization . 2021. WHO coronavirus disease (COVID-19) dashboard.https://covid19.who.int/ - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

