Targeting the glycans: A paradigm for host-targeted and COVID-19 drug design
- PMID: 34028178
- PMCID: PMC8242448
- DOI: 10.1111/jcmm.16585
Targeting the glycans: A paradigm for host-targeted and COVID-19 drug design
Abstract
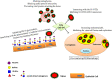
There is always a need for new approaches for the control of virus burdens caused by seasonal outbreaks, the emergence of novel viruses with pandemic potential and the development of resistance to current antiviral drugs. The outbreak of the 2019 novel coronavirus-disease COVID-19 represented a pandemic threat and declared a public health emergency of international concern. Herein, the role of glycans for the development of new drugs or vaccines, as a host-targeted approach, is discussed where may provide a front-line prophylactic or threats to protect against the current and any future respiratory-infecting virus and possibly against other respiratory pathogens. As a prototype, the role of glycans in the coronavirus infection, as well as, galectins (Gal) as the glycan-recognition agents (GRAs) in drug design are here summarized. Galectins, in particular, Gal-1 and Gal-3 are ubiquitous and important to biological systems, whose interactions with viral glycans modulate host immunity and homeostatic balance.
Keywords: coronavirus; glycan-recognition agents; glycans; spike protein.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Conflict of interest statement
There is no conflict of interest to be declared.
Figures



Similar articles
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
-
Problems associated with antiviral drugs and vaccines development for COVID-19: approach to intervention using expression vectors via GPI anchor.Nucleosides Nucleotides Nucleic Acids. 2021;40(6):665-706. doi: 10.1080/15257770.2021.1914851. Epub 2021 May 13. Nucleosides Nucleotides Nucleic Acids. 2021. PMID: 33982646 Free PMC article. Review.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Containing the spread of COVID-19 virus facing to its high mutation rate: approach to intervention using a nonspecific way of blocking its entry into the cells.Nucleosides Nucleotides Nucleic Acids. 2022;41(8):778-814. doi: 10.1080/15257770.2022.2071937. Epub 2022 May 9. Nucleosides Nucleotides Nucleic Acids. 2022. PMID: 35532338
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
Cited by
-
Seasonal effects decouple SARS-CoV-2 haplotypes worldwide.F1000Res. 2023 Mar 13;12:267. doi: 10.12688/f1000research.131522.1. eCollection 2023. F1000Res. 2023. PMID: 37069849 Free PMC article.
-
The emergence of SARS-CoV-2 variants of concern in Australia by haplotype coalescence reveals a continental link to COVID-19 seasonality.Methods Microbiol. 2022;50:233-268. doi: 10.1016/bs.mim.2022.03.003. Epub 2022 May 17. Methods Microbiol. 2022. PMID: 38013929 Free PMC article.
-
Blood Features Associated with Viral Infection Severity: An Experience from COVID-19-Pandemic Patients Hospitalized in the Center of Iran, Yazd.Int J Clin Pract. 2024 Mar 12;2024:7484645. doi: 10.1155/2024/7484645. eCollection 2024. Int J Clin Pract. 2024. PMID: 38505695 Free PMC article.
References
-
- Zhao X, Guo F, Comunale MA, et al. Inhibition of endoplasmic reticulum‐resident glucosidases impairs severe acute respiratory syndrome coronavirus and humancoronavirusNL63spikeprotein‐mediatedentry by altering the glycan processing of angiotensinI‐convertingenzyme2. Antimicrob Agents Chem other. 2015;59(1):206‐216. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

