Interfacing Seurat with the R tidy universe
- PMID: 34028547
- PMCID: PMC9502154
- DOI: 10.1093/bioinformatics/btab404
Interfacing Seurat with the R tidy universe
Abstract
Motivation: Seurat is one of the most popular software suites for the analysis of single-cell RNA sequencing data. Considering the popularity of the tidyverse ecosystem, which offers a large set of data display, query, manipulation, integration and visualization utilities, a great opportunity exists to interface the Seurat object with the tidyverse. This interface gives the large data science community of tidyverse users the possibility to operate with familiar grammar.
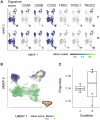
Results: To provide Seurat with a tidyverse-oriented interface without compromising efficiency, we developed tidyseurat, a lightweight adapter to the tidyverse. Tidyseurat displays cell information as a tibble abstraction, allowing intuitively interfacing Seurat with dplyr, tidyr, ggplot2 and plotly packages powering efficient data manipulation, integration and visualization. Iterative analyses on data subsets are enabled by interfacing with the popular nest-map framework.
Availability and implementation: The software is freely available at cran.r-project.org/web/packages/tidyseurat and github.com/stemangiola/tidyseurat.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures




References
-
- Bojanowski M., Edwards R. (2016) Alluvial: r package for creating alluvial diagrams. R Package Version 0. 1, 2.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

