Organization of the laminin polymer node
- PMID: 34029691
- PMCID: PMC8223250
- DOI: 10.1016/j.matbio.2021.05.004
Organization of the laminin polymer node
Abstract
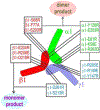
Laminin polymerization is a key step of basement membrane assembly that depends on the binding of α, β and γ N-terminal LN domains to form a polymer node. Nodal assembly can be divided into two steps consisting of β- and γ-LN dimerization followed by calcium-dependent addition of the α-LN domain. The assembly and structural organization of laminin-111 LN-LEa segments was examined by size-exclusion chromatography (SEC) and electron microscopy. Triskelion-like structures were observed in negatively-stained images of purified α1/β1/γ1 LN-LEa trimers. Image averaging of these revealed a heel-to-toe organization of the LN domains with angled outward projections of the LEa stem-like domains. A series of single-amino acid substitutions was introduced into the polymerization faces of the α1, β1 and γ1 LN domains followed by SEC analysis to distinguish between loss of β-γ mediated dimerization and loss of α-dependent trimerization (with intact β-γ dimers). Dimer-blocking mutations were confined to the γ1-toe and the β1-heel, whereas the trimer-only-blocking mutations mapped to the γ1-heel, β1-toe and the α1-toe and heel. Thus, in the polymer node the γ1-toe pairs with the β1-heel, the β1-toe pairs with the α1-heel, and the α1-toe pairs with the γ1-heel.
Keywords: Basement membrane; image averaging; ln mutations; self-assembly; triskelion.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

