Integrated Single-Cell Atlas of Endothelial Cells of the Human Lung
- PMID: 34030460
- PMCID: PMC8300155
- DOI: 10.1161/CIRCULATIONAHA.120.052318
Integrated Single-Cell Atlas of Endothelial Cells of the Human Lung
Abstract
Background: Cellular diversity of the lung endothelium has not been systematically characterized in humans. We provide a reference atlas of human lung endothelial cells (ECs) to facilitate a better understanding of the phenotypic diversity and composition of cells comprising the lung endothelium.
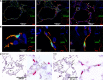
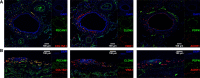
Methods: We reprocessed human control single-cell RNA sequencing (scRNAseq) data from 6 datasets. EC populations were characterized through iterative clustering with subsequent differential expression analysis. Marker genes were validated by fluorescent microscopy and in situ hybridization. scRNAseq of primary lung ECs cultured in vitro was performed. The signaling network between different lung cell types was studied. For cross-species analysis or disease relevance, we applied the same methods to scRNAseq data obtained from mouse lungs or from human lungs with pulmonary hypertension.
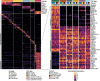
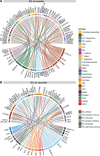
Results: Six lung scRNAseq datasets were reanalyzed and annotated to identify >15 000 vascular EC cells from 73 individuals. Differential expression analysis of EC revealed signatures corresponding to endothelial lineage, including panendothelial, panvascular, and subpopulation-specific marker gene sets. Beyond the broad cellular categories of lymphatic, capillary, arterial, and venous ECs, we found previously indistinguishable subpopulations; among venous EC, we identified 2 previously indistinguishable populations: pulmonary-venous ECs (COL15A1neg) localized to the lung parenchyma and systemic-venous ECs (COL15A1pos) localized to the airways and the visceral pleura; among capillary ECs, we confirmed their subclassification into recently discovered aerocytes characterized by EDNRB, SOSTDC1, and TBX2 and general capillary EC. We confirmed that all 6 endothelial cell types, including the systemic-venous ECs and aerocytes, are present in mice and identified endothelial marker genes conserved in humans and mice. Ligand-receptor connectome analysis revealed important homeostatic crosstalk of EC with other lung resident cell types. scRNAseq of commercially available primary lung ECs demonstrated a loss of their native lung phenotype in culture. scRNAseq revealed that endothelial diversity is maintained in pulmonary hypertension. Our article is accompanied by an online data mining tool (www.LungEndothelialCellAtlas.com).
Conclusions: Our integrated analysis provides a comprehensive and well-crafted reference atlas of ECs in the normal lung and confirms and describes in detail previously unrecognized endothelial populations across a large number of humans and mice.
Keywords: endothelial cells; microcirculation; pulmonary circulation; transcriptome.
Figures


References
-
- Katz AM. Knowledge of the circulation before William Harvey. Circulation. 1957; 15:726–734 - PubMed
-
- Vita JA. Endothelial function. Circulation. 2011; 124:e906–e912. doi: 10.1161/CIRCULATIONAHA.111.078824 - PubMed
-
- Deanfield JE, Halcox JP, Rabelink TJ. Endothelial function and dysfunction: testing and clinical relevance. Circulation. 2007; 115:1285–1295. doi: 10.1161/CIRCULATIONAHA.106.652859 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

