Transcription- and phosphorylation-dependent control of a functional interplay between XBP1s and PINK1 governs mitophagy and potentially impacts Parkinson disease pathophysiology
- PMID: 34030589
- PMCID: PMC8726674
- DOI: 10.1080/15548627.2021.1917129
Transcription- and phosphorylation-dependent control of a functional interplay between XBP1s and PINK1 governs mitophagy and potentially impacts Parkinson disease pathophysiology
Abstract
Parkinson disease (PD)-affected brains show consistent endoplasmic reticulum (ER) stress and mitophagic dysfunctions. The mechanisms underlying these perturbations and how they are directly linked remain a matter of questions. XBP1 is a transcription factor activated upon ER stress after unconventional splicing by the nuclease ERN1/IREα thereby yielding XBP1s, whereas PINK1 is a kinase considered as the sensor of mitochondrial physiology and a master gatekeeper of mitophagy process. We showed that XBP1s transactivates PINK1 in human cells, primary cultured neurons and mice brain, and triggered a pro-mitophagic phenotype that was fully dependent of endogenous PINK1. We also unraveled a PINK1-dependent phosphorylation of XBP1s that conditioned its nuclear localization and thereby, governed its transcriptional activity. PINK1-induced XBP1s phosphorylation occurred at residues reminiscent of, and correlated to, those phosphorylated in substantia nigra of sporadic PD-affected brains. Overall, our study delineated a functional loop between XBP1s and PINK1 governing mitophagy that was disrupted in PD condition.Abbreviations: 6OHDA: 6-hydroxydopamine; baf: bafilomycin A1; BECN1: beclin 1; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CASP3: caspase 3; CCCP: carbonyl cyanide chlorophenylhydrazone; COX8A: cytochrome c oxidase subunit 8A; DDIT3/CHOP: DNA damage inducible transcript 3; EGFP: enhanced green fluorescent protein; ER: endoplasmic reticulum; ERN1/IRE1α: endoplasmic reticulum to nucleus signaling 1; FACS: fluorescence-activated cell sorting; HSPD1/HSP60: heat shock protein family D (Hsp60) member 1; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MFN2: mitofusin 2; OPTN: optineurin; PD: Parkinson disease; PINK1: PTEN-induced kinase 1; PCR: polymerase chain reaction:; PRKN: parkin RBR E3 ubiquitin protein ligase; XBP1s [p-S61A]: XBP1s phosphorylated at serine 61; XBP1s [p-T48A]: XBP1s phosphorylated at threonine 48; shRNA: short hairpin RNA, SQSTM1/p62: sequestosome 1; TIMM23: translocase of inner mitochondrial membrane 23; TM: tunicamycin; TMRM: tetramethyl rhodamine methylester; TOMM20: translocase of outer mitochondrial membrane 20; Toy: toyocamycin; TP: thapsigargin; UB: ubiquitin; UB (S65): ubiquitin phosphorylated at serine 65; UPR: unfolded protein response, XBP1: X-box binding protein 1; XBP1s: spliced X-box binding protein 1.
Keywords: Mitophagy; PINK1; Parkinson disease; XBP1; phosphorylation; transcription; unfolded protein response.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


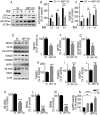
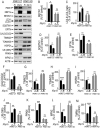

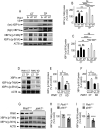
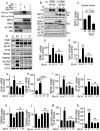
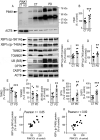
References
-
- Sugeno N, Takeda A, Hasegawa T, et al. Serine 129 phosphorylation of alpha-synuclein induces unfolded protein response-mediated cell death. J Biol Chem. 2008;283:23179–23188. - PubMed
-
- Hetz C, Saxena S.. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol. 2017;13:477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
