The metabolic profile of Bifidobacterium dentium reflects its status as a human gut commensal
- PMID: 34030655
- PMCID: PMC8145834
- DOI: 10.1186/s12866-021-02166-6
The metabolic profile of Bifidobacterium dentium reflects its status as a human gut commensal
Abstract
Background: Bifidobacteria are commensal microbes of the mammalian gastrointestinal tract. In this study, we aimed to identify the intestinal colonization mechanisms and key metabolic pathways implemented by Bifidobacterium dentium.
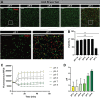
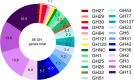
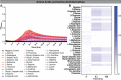
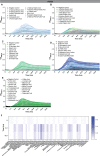
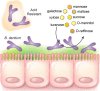
Results: B. dentium displayed acid resistance, with high viability over a pH range from 4 to 7; findings that correlated to the expression of Na+/H+ antiporters within the B. dentium genome. B. dentium was found to adhere to human MUC2+ mucus and harbor mucin-binding proteins. Using microbial phenotyping microarrays and fully-defined media, we demonstrated that in the absence of glucose, B. dentium could metabolize a variety of nutrient sources. Many of these nutrient sources were plant-based, suggesting that B. dentium can consume dietary substances. In contrast to other bifidobacteria, B. dentium was largely unable to grow on compounds found in human mucus; a finding that was supported by its glycosyl hydrolase (GH) profile. Of the proteins identified in B. dentium by proteomic analysis, a large cohort of proteins were associated with diverse metabolic pathways, indicating metabolic plasticity which supports colonization of the dynamic gastrointestinal environment.
Conclusions: Taken together, we conclude that B. dentium is well adapted for commensalism in the gastrointestinal tract.
Keywords: Acid stress; Bifidobacteria; Carbohydrates; Commensal; Glycans; Intestine; Metabolism.
Conflict of interest statement
J. Versalovic serves on the scientific advisory boards of Biomica, Plexus Worldwide and Seed Health. R.A. Britton serves on the scientific advisory board of Tenza, is a co-founder of Mikrovia, and consults for Takeda and Probiotech. J. Versalovic, J.K. Spinler, and R.A. Britton have received unrestricted research support from BioGaia, AB. The remaining authors have no commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways.mBio. 2019 Jun 18;10(3):e01087-19. doi: 10.1128/mBio.01087-19. mBio. 2019. PMID: 31213556 Free PMC article.
-
The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity.PLoS Genet. 2009 Dec;5(12):e1000785. doi: 10.1371/journal.pgen.1000785. Epub 2009 Dec 24. PLoS Genet. 2009. PMID: 20041198 Free PMC article.
-
Bifidobacterium dentium-derived y-glutamylcysteine suppresses ER-mediated goblet cell stress and reduces TNBS-driven colonic inflammation.Gut Microbes. 2021 Jan-Dec;13(1):1-21. doi: 10.1080/19490976.2021.1902717. Gut Microbes. 2021. PMID: 33985416 Free PMC article.
-
Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health.Appl Microbiol Biotechnol. 2019 Sep;103(18):7287-7315. doi: 10.1007/s00253-019-10012-z. Epub 2019 Jul 22. Appl Microbiol Biotechnol. 2019. PMID: 31332487 Review.
-
Genomics and ecological overview of the genus Bifidobacterium.Int J Food Microbiol. 2011 Sep 1;149(1):37-44. doi: 10.1016/j.ijfoodmicro.2010.12.010. Epub 2010 Dec 28. Int J Food Microbiol. 2011. PMID: 21276626 Review.
Cited by
-
Synergistic Effects of Probiotics and Lifestyle Interventions on Intestinal Microbiota Composition and Clinical Outcomes in Obese Adults.Metabolites. 2025 Jan 23;15(2):70. doi: 10.3390/metabo15020070. Metabolites. 2025. PMID: 39997695 Free PMC article.
-
Unraveling the Metabolic Requirements of the Gut Commensal Bacteroides ovatus.Front Microbiol. 2021 Nov 25;12:745469. doi: 10.3389/fmicb.2021.745469. eCollection 2021. Front Microbiol. 2021. PMID: 34899632 Free PMC article.
-
Diversity Analysis of Intestinal Bifidobacteria in the Hohhot Population.Microorganisms. 2024 Apr 9;12(4):756. doi: 10.3390/microorganisms12040756. Microorganisms. 2024. PMID: 38674700 Free PMC article.
-
Identifying single-strain growth patterns of human gut microbes in response to preterm human milk and formula.Food Funct. 2022 May 23;13(10):5571-5589. doi: 10.1039/d2fo00447j. Food Funct. 2022. PMID: 35481924 Free PMC article.
-
Development of gut microbiota during the first 2 years of life.Sci Rep. 2022 May 31;12(1):9080. doi: 10.1038/s41598-022-13009-3. Sci Rep. 2022. PMID: 35641542 Free PMC article.
References
-
- He M, Li M, Wang SY, Zhang LL, Miao JJ, Shi L, Yu Q, Yao JR, Huang CY, He F. Analyzing colonization of Bifidobacteria in infants with real-time fluorescent quantitative PCR. Sichuan Da Xue Xue Bao Yi Xue Ban. 2016;47(4):527–532. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

