The epigenetic regulator G9a attenuates stress-induced resistance and metabolic transcriptional programs across different stressors and species
- PMID: 34030685
- PMCID: PMC8142638
- DOI: 10.1186/s12915-021-01025-0
The epigenetic regulator G9a attenuates stress-induced resistance and metabolic transcriptional programs across different stressors and species
Abstract
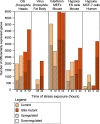
Background: Resistance and tolerance are two coexisting defense strategies for fighting infections. Resistance is mediated by signaling pathways that induce transcriptional activation of resistance factors that directly eliminate the pathogen. Tolerance refers to adaptations that limit the health impact of a given pathogen burden, without targeting the infectious agent. The key players governing immune tolerance are largely unknown. In Drosophila, the histone H3 lysine 9 (H3K9) methyltransferase G9a was shown to mediate tolerance to virus infection and oxidative stress (OS), suggesting that abiotic stresses like OS may also evoke tolerance mechanisms. In response to both virus and OS, stress resistance genes were overinduced in Drosophila G9a mutants, suggesting an intact but overactive stress response. We recently demonstrated that G9a promotes tolerance to OS by maintaining metabolic homeostasis and safeguarding energy availability, but it remained unclear if this mechanism also applies to viral infection, or is conserved in other species and stress responses. To address these questions, we analyzed publicly available datasets from Drosophila, mouse, and human in which global gene expression levels were measured in G9a-depleted conditions and controls at different time points upon stress exposure.
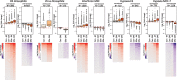
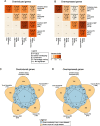
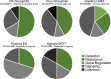
Results: In all investigated datasets, G9a attenuates the transcriptional stress responses that confer resistance against the encountered stressor. Comparative analysis of conserved G9a-dependent stress response genes suggests that G9a is an intimate part of the design principles of stress resistance, buffering the induction of promiscuous stress signaling pathways and stress-specific resistance factors. Importantly, we find stress-dependent downregulation of metabolic genes to also be dependent on G9a across all of the tested datasets.
Conclusions: These results suggest that G9a sets the balance between activation of resistance genes and maintaining metabolic homeostasis, thereby ensuring optimal organismal performance during exposure to diverse types of stress across different species. We therefore propose G9a as a potentially conserved master regulator underlying the widely important, yet poorly understood, concept of stress tolerance.
Keywords: Drosophila; G9a; Mammalian cells; Metabolism; Resistance; Stress response; Tolerance; Transcription.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
The histone methyltransferase G9a regulates tolerance to oxidative stress-induced energy consumption.PLoS Biol. 2019 Mar 12;17(3):e2006146. doi: 10.1371/journal.pbio.2006146. eCollection 2019 Mar. PLoS Biol. 2019. PMID: 30860988 Free PMC article.
-
Epigenetic regulation of starvation-induced autophagy in Drosophila by histone methyltransferase G9a.Sci Rep. 2017 Aug 4;7(1):7343. doi: 10.1038/s41598-017-07566-1. Sci Rep. 2017. PMID: 28779125 Free PMC article.
-
Histone Methyltransferase G9a Is Required for Cardiomyocyte Homeostasis and Hypertrophy.Circulation. 2017 Sep 26;136(13):1233-1246. doi: 10.1161/CIRCULATIONAHA.117.028561. Epub 2017 Aug 4. Circulation. 2017. PMID: 28778944
-
H3K9 methyltransferase G9a and the related molecule GLP.Genes Dev. 2011 Apr 15;25(8):781-8. doi: 10.1101/gad.2027411. Genes Dev. 2011. PMID: 21498567 Free PMC article. Review.
-
Targeting EHMT2/ G9a for cancer therapy: Progress and perspective.Eur J Pharmacol. 2021 Feb 15;893:173827. doi: 10.1016/j.ejphar.2020.173827. Epub 2020 Dec 19. Eur J Pharmacol. 2021. PMID: 33347828
Cited by
-
Intraspecific genetic variation in host vigour, viral load and disease tolerance during Drosophila C virus infection.Open Biol. 2023 Mar;13(3):230025. doi: 10.1098/rsob.230025. Epub 2023 Mar 1. Open Biol. 2023. PMID: 36854375 Free PMC article.
-
Regulation of paternal 5mC oxidation and H3K9me2 asymmetry by ERK1/2 in mouse zygotes.Cell Biosci. 2022 Mar 7;12(1):25. doi: 10.1186/s13578-022-00758-x. Cell Biosci. 2022. PMID: 35255956 Free PMC article.
-
SetDB1 and Su(var)3-9 are essential for late stages of larval development of Drosophila melanogaster.Chromosome Res. 2023 Dec 15;31(4):35. doi: 10.1007/s10577-023-09743-7. Chromosome Res. 2023. PMID: 38099968
-
Growth, body composition, and endocrine-metabolic profiles of individuals with Kleefstra syndrome provide directions for clinical management and translational studies.Am J Med Genet A. 2024 May;194(5):e63472. doi: 10.1002/ajmg.a.63472. Epub 2023 Dec 29. Am J Med Genet A. 2024. PMID: 38155610 Free PMC article.
-
Epigenetic responses in Borrelia-infected Ixodes scapularis ticks: Over-expression of euchromatic histone lysine methyltransferase 2 and no change in DNA methylation.PLoS One. 2025 Jun 5;20(6):e0324546. doi: 10.1371/journal.pone.0324546. eCollection 2025. PLoS One. 2025. PMID: 40472016 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

