Immunotranscriptomic profiling the acute and clearance phases of a human challenge dengue virus serotype 2 infection model
- PMID: 34031380
- PMCID: PMC8144425
- DOI: 10.1038/s41467-021-22930-6
Immunotranscriptomic profiling the acute and clearance phases of a human challenge dengue virus serotype 2 infection model
Abstract
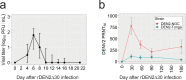
About 20-25% of dengue virus (DENV) infections become symptomatic ranging from self-limiting fever to shock. Immune gene expression changes during progression to severe dengue have been documented in hospitalized patients; however, baseline or kinetic information is difficult to standardize in natural infection. Here we profile the host immunotranscriptome response in humans before, during, and after infection with a partially attenuated rDEN2Δ30 challenge virus (ClinicalTrials.gov NCT02021968). Inflammatory genes including type I interferon and viral restriction pathways are induced during DENV2 viremia and return to baseline after viral clearance, while others including myeloid, migratory, humoral, and growth factor immune regulation factors pathways are found at non-baseline levels post-viremia. Furthermore, pre-infection baseline gene expression is useful to predict rDEN2Δ30-induced immune responses and the development of rash. Our results suggest a distinct immunological profile for mild rDEN2Δ30 infection and offer new potential biomarkers for characterizing primary DENV infection.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- WHO. Dengue guidelines for diagnosis, treatment, prevention and control: new edition.) World Health Organization (2009). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

