Identification of microbial markers across populations in early detection of colorectal cancer
- PMID: 34031391
- PMCID: PMC8144394
- DOI: 10.1038/s41467-021-23265-y
Identification of microbial markers across populations in early detection of colorectal cancer
Abstract
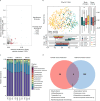
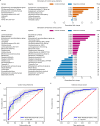
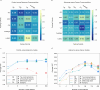
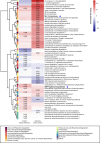
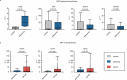
Associations between gut microbiota and colorectal cancer (CRC) have been widely investigated. However, the replicable markers for early-stage adenoma diagnosis across multiple populations remain elusive. Here, we perform an integrated analysis on 1056 public fecal samples, to identify adenoma-associated microbial markers for early detection of CRC. After adjusting for potential confounders, Random Forest classifiers are constructed with 11 markers to discriminate adenoma from control (area under the ROC curve (AUC) = 0.80), and 26 markers to discriminate adenoma from CRC (AUC = 0.89), respectively. Moreover, we validate the classifiers in two independent cohorts achieving AUCs of 0.78 and 0.84, respectively. Functional analysis reveals that the altered microbiome is characterized with increased ADP-L-glycero-beta-D-manno-heptose biosynthesis in adenoma and elevated menaquinone-10 biosynthesis in CRC. These findings are validated in a newly-collected cohort of 43 samples using quantitative real-time PCR. This work proves the validity of adenoma-specific markers across multi-populations, which would contribute to the early diagnosis and treatment of CRC.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

