CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data
- PMID: 34031394
- PMCID: PMC8144550
- DOI: 10.1038/s41467-021-22417-4
CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data
Abstract
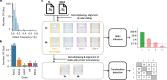
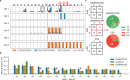
Controlling off-target editing activity is one of the central challenges in making CRISPR technology accurate and applicable in medical practice. Current algorithms for analyzing off-target activity do not provide statistical quantification, are not sufficiently sensitive in separating signal from noise in experiments with low editing rates, and do not address the detection of translocations. Here we present CRISPECTOR, a software tool that supports the detection and quantification of on- and off-target genome-editing activity from NGS data using paired treatment/control CRISPR experiments. In particular, CRISPECTOR facilitates the statistical analysis of NGS data from multiplex-PCR comparative experiments to detect and quantify adverse translocation events. We validate the observed results and show independent evidence of the occurrence of translocations in human cell lines, after genome editing. Our methodology is based on a statistical model comparison approach leading to better false-negative rates in sites with weak yet significant off-target activity.
Conflict of interest statement
G.K., M.S.M., and G.R.R. are employees of Integrated DNA Technologies (IDT), which sells reagents similar to some described in the manuscript. All other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

