Fast widefield scan provides tunable and uniform illumination optimizing super-resolution microscopy on large fields
- PMID: 34031402
- PMCID: PMC8144377
- DOI: 10.1038/s41467-021-23405-4
Fast widefield scan provides tunable and uniform illumination optimizing super-resolution microscopy on large fields
Abstract
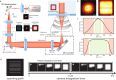
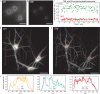
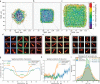
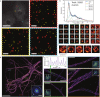
Non-uniform illumination limits quantitative analyses of fluorescence imaging techniques. In particular, single molecule localization microscopy (SMLM) relies on high irradiances, but conventional Gaussian-shaped laser illumination restricts the usable field of view to around 40 µm × 40 µm. We present Adaptable Scanning for Tunable Excitation Regions (ASTER), a versatile illumination technique that generates uniform and adaptable illumination. ASTER is also highly compatible with optical sectioning techniques such as total internal reflection fluorescence (TIRF). For SMLM, ASTER delivers homogeneous blinking kinetics at reasonable laser power over fields-of-view up to 200 µm × 200 µm. We demonstrate that ASTER improves clustering analysis and nanoscopic size measurements by imaging nanorulers, microtubules and clathrin-coated pits in COS-7 cells, and β2-spectrin in neurons. ASTER's sharp and quantitative illumination paves the way for high-throughput quantification of biological structures and processes in classical and super-resolution fluorescence microscopies.
Conflict of interest statement
N.B. and S.L.F. are shareholders in Abbelight. Other authors declare no competing interests.
Figures

Similar articles
-
Imaging cellular structures in super-resolution with SIM, STED and Localisation Microscopy: A practical comparison.Sci Rep. 2016 Jun 6;6:27290. doi: 10.1038/srep27290. Sci Rep. 2016. PMID: 27264341 Free PMC article.
-
Homogeneous multifocal excitation for high-throughput super-resolution imaging.Nat Methods. 2020 Jul;17(7):726-733. doi: 10.1038/s41592-020-0859-z. Epub 2020 Jun 22. Nat Methods. 2020. PMID: 32572233
-
Spinning-Spot Shadowless TIRF Microscopy.PLoS One. 2015 Aug 26;10(8):e0136055. doi: 10.1371/journal.pone.0136055. eCollection 2015. PLoS One. 2015. PMID: 26308212 Free PMC article.
-
About samples, giving examples: Optimized Single Molecule Localization Microscopy.Methods. 2020 Mar 1;174:100-114. doi: 10.1016/j.ymeth.2019.05.008. Epub 2019 May 10. Methods. 2020. PMID: 31078795 Review.
-
Spectroscopic analysis beyond the diffraction limit.Int J Biochem Cell Biol. 2018 Aug;101:113-117. doi: 10.1016/j.biocel.2018.06.002. Epub 2018 Jun 3. Int J Biochem Cell Biol. 2018. PMID: 29874548 Free PMC article. Review.
Cited by
-
Correlative single-molecule and structured illumination microscopy of fast dynamics at the plasma membrane.Nat Commun. 2024 Jul 10;15(1):5813. doi: 10.1038/s41467-024-49876-9. Nat Commun. 2024. PMID: 38987559 Free PMC article.
-
Fluorescence nanoscopy at the sub-10 nm scale.Biophys Rev. 2021 Dec 2;13(6):1101-1112. doi: 10.1007/s12551-021-00864-z. eCollection 2021 Dec. Biophys Rev. 2021. PMID: 35059030 Free PMC article. Review.
-
An open-source microscopy framework for simultaneous control of image acquisition, reconstruction, and analysis.HardwareX. 2023 Feb 6;13:e00400. doi: 10.1016/j.ohx.2023.e00400. eCollection 2023 Mar. HardwareX. 2023. PMID: 36824447 Free PMC article.
-
PCIe-based FPGA-GPU heterogeneous computation for real-time multi-emitter fitting in super-resolution localization microscopy.Biomed Opt Express. 2022 May 16;13(6):3401-3415. doi: 10.1364/BOE.459198. eCollection 2022 Jun 1. Biomed Opt Express. 2022. PMID: 35781968 Free PMC article.
-
A new mechanism of fibronectin fibril assembly revealed by live imaging and super-resolution microscopy.J Cell Sci. 2022 Aug 15;135(16):jcs260120. doi: 10.1242/jcs.260120. Epub 2022 Aug 25. J Cell Sci. 2022. PMID: 35851804 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

