hTERT-immortalized gingival fibroblasts respond to cytokines but fail to mimic primary cell responses to Porphyromonas gingivalis
- PMID: 34031466
- PMCID: PMC8144196
- DOI: 10.1038/s41598-021-90037-5
hTERT-immortalized gingival fibroblasts respond to cytokines but fail to mimic primary cell responses to Porphyromonas gingivalis
Abstract
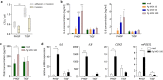
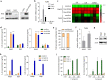
In periodontitis, gingival fibroblasts (GFs) interact with and respond to oral pathogens, significantly contributing to perpetuation of chronic inflammation and tissue destruction. The aim of this study was to determine the usefulness of the recently released hTERT-immortalized GF (TIGF) cell line for studies of host-pathogen interactions. We show that TIGFs are unable to upregulate expression and production of interleukin (IL)-6, IL-8 and prostaglandin E2 upon infection with Porphyromonas gingivalis despite being susceptible to adhesion and invasion by this oral pathogen. In contrast, induction of inflammatory mediators in TNFα- or IL-1β-stimulated TIGFs is comparable to that observed in primary GFs. The inability of TIGFs to respond directly to P. gingivalis is caused by a specific defect in Toll-like receptor-2 (TLR2) expression, which is likely driven by TLR2 promoter hypermethylation. Consistently, TIGFs fail to upregulate inflammatory genes in response to the TLR2 agonists Pam2CSK4 and Pam3CSK4. These results identify important limitations of using TIGFs to study GF interaction with oral pathogens, though these cells may be useful for studies of TLR2-independent processes. Our observations also emphasize the importance of direct comparisons between immortalized and primary cells prior to using cell lines as models in studies of any biological processes.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Synergistic induction of PGE2 by oral pathogens and TNF promotes gingival fibroblast-driven stromal-immune cross-talk in periodontitis.mBio. 2025 May 14;16(5):e0004625. doi: 10.1128/mbio.00046-25. Epub 2025 Apr 3. mBio. 2025. PMID: 40178270 Free PMC article.
-
TLR2 promoter hypermethylation creates innate immune dysbiosis.J Dent Res. 2015 Jan;94(1):183-91. doi: 10.1177/0022034514557545. Epub 2014 Nov 11. J Dent Res. 2015. PMID: 25389002 Free PMC article.
-
Chronic Exposure of Gingival Fibroblasts to TLR2 or TLR4 Agonist Inhibits Osteoclastogenesis but Does Not Affect Osteogenesis.Front Immunol. 2020 Jul 23;11:1693. doi: 10.3389/fimmu.2020.01693. eCollection 2020. Front Immunol. 2020. PMID: 32793243 Free PMC article.
-
Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts-CD14 and Toll-like receptors.Crit Rev Oral Biol Med. 2002;13(2):132-42. doi: 10.1177/154411130201300204. Crit Rev Oral Biol Med. 2002. PMID: 12097356 Review.
-
The Role of Gingival Fibroblasts in the Pathogenesis of Periodontitis.J Dent Res. 2023 May;102(5):489-496. doi: 10.1177/00220345231151921. Epub 2023 Mar 8. J Dent Res. 2023. PMID: 36883660 Free PMC article. Review.
Cited by
-
Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies.Dent J (Basel). 2023 Jun 26;11(7):158. doi: 10.3390/dj11070158. Dent J (Basel). 2023. PMID: 37504224 Free PMC article. Review.
-
TLR2 Activation by Porphyromonas gingivalis Requires Both PPAD Activity and Fimbriae.Front Immunol. 2022 Apr 1;13:823685. doi: 10.3389/fimmu.2022.823685. eCollection 2022. Front Immunol. 2022. PMID: 35432342 Free PMC article.
-
Matrix Metalloproteinases in the Periodontium-Vital in Tissue Turnover and Unfortunate in Periodontitis.Int J Mol Sci. 2024 Feb 27;25(5):2763. doi: 10.3390/ijms25052763. Int J Mol Sci. 2024. PMID: 38474009 Free PMC article. Review.
-
Three-Dimensional Humanized Model of the Periodontal Gingival Pocket to Study Oral Microbiome.Adv Sci (Weinh). 2023 Apr;10(12):e2205473. doi: 10.1002/advs.202205473. Epub 2023 Feb 24. Adv Sci (Weinh). 2023. PMID: 36825685 Free PMC article.
-
Molecular Research on Oral Diseases and Related Biomaterials: A Journey from Oral Cell Models to Advanced Regenerative Perspectives.Int J Mol Sci. 2022 May 9;23(9):5288. doi: 10.3390/ijms23095288. Int J Mol Sci. 2022. PMID: 35563679 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

