CD13 is a critical regulator of cell-cell fusion in osteoclastogenesis
- PMID: 34031489
- PMCID: PMC8144195
- DOI: 10.1038/s41598-021-90271-x
CD13 is a critical regulator of cell-cell fusion in osteoclastogenesis
Abstract
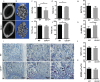
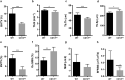
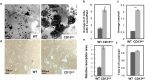
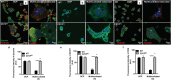
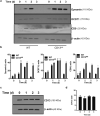
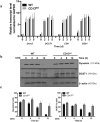
The transmembrane aminopeptidase CD13 is highly expressed in cells of the myeloid lineage, regulates dynamin-dependent receptor endocytosis and recycling and is a necessary component of actin cytoskeletal organization. Here, we show that CD13-deficient mice present a low bone density phenotype with increased numbers of osteoclasts per bone surface, but display a normal distribution of osteoclast progenitor populations in the bone marrow and periphery. In addition, the bone formation and mineral apposition rates are similar between genotypes, indicating a defect in osteoclast-specific function in vivo. Lack of CD13 led to exaggerated in vitro osteoclastogenesis as indicated by significantly enhanced fusion of bone marrow-derived multinucleated osteoclasts in the presence of M-CSF and RANKL, resulting in abnormally large cells containing remarkably high numbers of nuclei. Mechanistically, while expression levels of the fusion-regulatory proteins dynamin and DC-STAMP1 must be downregulated for fusion to proceed, these are aberrantly sustained at high levels even in CD13-deficient mature multi-nucleated osteoclasts. Further, the stability of fusion-promoting proteins is maintained in the absence of CD13, implicating CD13 in protein turnover mechanisms. Together, we conclude that CD13 may regulate cell-cell fusion by controlling the expression and localization of key fusion regulatory proteins that are critical for osteoclast fusion.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Low-density lipoprotein receptor deficiency causes impaired osteoclastogenesis and increased bone mass in mice because of defect in osteoclastic cell-cell fusion.J Biol Chem. 2012 Jun 1;287(23):19229-41. doi: 10.1074/jbc.M111.323600. Epub 2012 Apr 12. J Biol Chem. 2012. PMID: 22500026 Free PMC article.
-
Targeted disruption of ephrin B1 in cells of myeloid lineage increases osteoclast differentiation and bone resorption in mice.PLoS One. 2012;7(3):e32887. doi: 10.1371/journal.pone.0032887. Epub 2012 Mar 5. PLoS One. 2012. PMID: 22403721 Free PMC article.
-
RARγ is a negative regulator of osteoclastogenesis.J Steroid Biochem Mol Biol. 2015 Jun;150:46-53. doi: 10.1016/j.jsbmb.2015.03.005. Epub 2015 Mar 20. J Steroid Biochem Mol Biol. 2015. PMID: 25800721
-
Regulators of osteoclast differentiation and cell-cell fusion.Keio J Med. 2011;60(4):101-5. doi: 10.2302/kjm.60.101. Keio J Med. 2011. PMID: 22200633 Review.
-
The dendritic cell-specific transmembrane protein DC-STAMP is essential for osteoclast fusion and osteoclast bone-resorbing activity.Mod Rheumatol. 2006;16(6):341-2. doi: 10.1007/s10165-006-0524-0. Epub 2006 Dec 20. Mod Rheumatol. 2006. PMID: 17164993 Review.
Cited by
-
Gastroesophageal circulating tumor cell crosstalk with peripheral immune system guides CTC survival and proliferation.Cell Death Dis. 2025 Mar 29;16(1):223. doi: 10.1038/s41419-025-07530-2. Cell Death Dis. 2025. PMID: 40157906 Free PMC article.
-
The Role of the Ectopeptidase APN/CD13 in Cancer.Biomedicines. 2023 Feb 28;11(3):724. doi: 10.3390/biomedicines11030724. Biomedicines. 2023. PMID: 36979703 Free PMC article. Review.
-
The Implant-Induced Foreign Body Response Is Limited by CD13-Dependent Regulation of Ubiquitination of Fusogenic Proteins.J Immunol. 2024 Feb 15;212(4):663-676. doi: 10.4049/jimmunol.2300688. J Immunol. 2024. PMID: 38149920 Free PMC article.
-
Long noncoding RNA Malat1 protects against osteoporosis and bone metastasis.Nat Commun. 2024 Mar 16;15(1):2384. doi: 10.1038/s41467-024-46602-3. Nat Commun. 2024. PMID: 38493144 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

