SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome
- PMID: 34031561
- PMCID: PMC8249329
- DOI: 10.1038/s41557-021-00707-0
SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome
Abstract
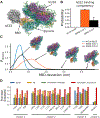
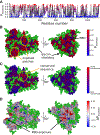
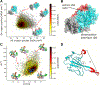
SARS-CoV-2 has intricate mechanisms for initiating infection, immune evasion/suppression and replication that depend on the structure and dynamics of its constituent proteins. Many protein structures have been solved, but far less is known about their relevant conformational changes. To address this challenge, over a million citizen scientists banded together through the Folding@home distributed computing project to create the first exascale computer and simulate 0.1 seconds of the viral proteome. Our adaptive sampling simulations predict dramatic opening of the apo spike complex, far beyond that seen experimentally, explaining and predicting the existence of 'cryptic' epitopes. Different spike variants modulate the probabilities of open versus closed structures, balancing receptor binding and immune evasion. We also discover dramatic conformational changes across the proteome, which reveal over 50 'cryptic' pockets that expand targeting options for the design of antivirals. All data and models are freely available online, providing a quantitative structural atlas.
Figures

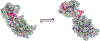
Update of
-
SARS-CoV-2 Simulations Go Exascale to Capture Spike Opening and Reveal Cryptic Pockets Across the Proteome.bioRxiv [Preprint]. 2020 Oct 7:2020.06.27.175430. doi: 10.1101/2020.06.27.175430. bioRxiv. 2020. Update in: Nat Chem. 2021 Jul;13(7):651-659. doi: 10.1038/s41557-021-00707-0. PMID: 32637963 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

