This is a preprint.
SARS-CoV-2 Infection Causes Dopaminergic Neuron Senescence
- PMID: 34031650
- PMCID: PMC8142658
- DOI: 10.21203/rs.3.rs-513461/v1
SARS-CoV-2 Infection Causes Dopaminergic Neuron Senescence
Update in
-
SARS-CoV-2 infection causes dopaminergic neuron senescence.Cell Stem Cell. 2024 Feb 1;31(2):196-211.e6. doi: 10.1016/j.stem.2023.12.012. Epub 2024 Jan 17. Cell Stem Cell. 2024. PMID: 38237586 Free PMC article.
Abstract
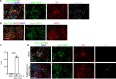
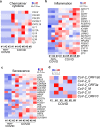
COVID-19 patients commonly present with neurological signs of central nervous system (CNS)1-3 and/or peripheral nervous system dysfunction4. However, which neural cells are permissive to infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been controversial. Here, we show that midbrain dopamine (DA) neurons derived from human pluripotent stem cells (hPSCs) are selectively permissive to SARS-CoV-2 infection both in vitro and upon transplantation in vivo, and that SARS-CoV-2 infection triggers a DA neuron inflammatory and cellular senescence response. A high-throughput screen in hPSC-derived DA neurons identified several FDA approved drugs, including riluzole, metformin, and imatinib, that can rescue the cellular senescence phenotype and prevent SARS-CoV-2 infection. RNA-seq analysis of human ventral midbrain tissue from COVID-19 patients, using formalin-fixed paraffin-embedded autopsy samples, confirmed the induction of an inflammatory and cellular senescence signature and identified low levels of SARS-CoV-2 transcripts. Our findings demonstrate that hPSC-derived DA neurons can serve as a disease model to study neuronal susceptibility to SARS-CoV-2 and to identify candidate neuroprotective drugs for COVID-19 patients. The susceptibility of hPSC-derived DA neurons to SARS-CoV-2 and the observed inflammatory and senescence transcriptional responses suggest the need for careful, long-term monitoring of neurological problems in COVID-19 patients.
Keywords: SARS-CoV-2; midbrain dopamine; neuron senescence.
Conflict of interest statement
Competing Interests. R.E.S. is on the scientific advisory board of Miromatrix Inc and is a paid consultant and speaker for Alnylam Inc. L.S. is a scientific cofounder and paid consultant of BlueRock Therapeutics Inc. The other authors declare no competing interests.
Figures



References
Publication types
Grants and funding
- DP3 DK111907/DK/NIDDK NIH HHS/United States
- R01 NS117583/NS/NINDS NIH HHS/United States
- R01 NS107442/NS/NINDS NIH HHS/United States
- R21 NS111176/NS/NINDS NIH HHS/United States
- R01 CA234614/CA/NCI NIH HHS/United States
- R01 DK124463/DK/NIDDK NIH HHS/United States
- F32 HD096810/HD/NICHD NIH HHS/United States
- R03 DK117252/DK/NIDDK NIH HHS/United States
- R01 DK116075/DK/NIDDK NIH HHS/United States
- R01 DK121072/DK/NIDDK NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
- R01 NS099270/NS/NINDS NIH HHS/United States
- R21 AG064596/AG/NIA NIH HHS/United States
- R01 DK119667/DK/NIDDK NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

