This is a preprint.
Durability of mRNA-1273-induced antibodies against SARS-CoV-2 variants
- PMID: 34031659
- PMCID: PMC8142657
- DOI: 10.1101/2021.05.13.444010
Durability of mRNA-1273-induced antibodies against SARS-CoV-2 variants
Update in
-
Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants.Science. 2021 Sep 17;373(6561):1372-1377. doi: 10.1126/science.abj4176. Epub 2021 Aug 13. Science. 2021. PMID: 34385356 Free PMC article.
Abstract
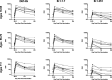
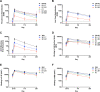
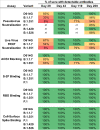
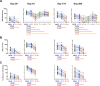
SARS-CoV-2 mutations may diminish vaccine-induced protective immune responses, and the durability of such responses has not been previously reported. Here, we present a comprehensive assessment of the impact of variants B.1.1.7, B.1.351, P.1, B.1.429, and B.1.526 on binding, neutralizing, and ACE2-blocking antibodies elicited by the vaccine mRNA-1273 over seven months. Cross-reactive neutralizing responses were rare after a single dose of mRNA-1273. At the peak of response to the second dose, all subjects had robust responses to all variants. Binding and functional antibodies against variants persisted in most subjects, albeit at low levels, for 6 months after the primary series of mRNA-1273. Across all assays, B.1.351 had the greatest impact on antibody recognition, and B.1.1.7 the least. These data complement ongoing studies of clinical protection to inform the potential need for additional boost vaccinations.
One-sentence summary: Most mRNA-1273 vaccinated individuals maintained binding and functional antibodies against SARS-CoV-2 variants for 6 months.
Conflict of interest statement
Competing interests:
Ms. Bennett and Dr. Leav are employees of Moderna, Inc.
Dr. Graham reports having a patent, International Patent Application No. WO/2018/081318 entitled “Prefusion Coronavirus Spike Proteins and Their Use” pending, and a patent, US Patent Application No. 62/972,886 entitled “2019-nCoV Vaccine” pending.
Dr. Anderson reports grants and personal fees from Pfizer, grants from Merck, grants from PaxVax, grants from Micron, grants and personal fees from Sanofi-Pasteur, grants from Janssen, grants from MedImmune, grants from GSK, personal fees from Medscape, personal fees from Kentucky Bioprocessing, Inc, outside the submitted work.
Dr. Rouphael reports grants from Pfizer, grants from Merck, grants from Sanofi-Pasteur, grants from Eli Lilly, grants from Quidel, outside the submitted work.
Figures

References
-
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). 2021. (https://coronavirus.jhu.edu/map.html ).
-
- Wibmer C. K. et al., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27, 622–625 (2021). - PubMed
-
- Wang P. et al., Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
