Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer's disease screening and staging
- PMID: 34032364
- PMCID: PMC9292367
- DOI: 10.1002/alz.12369
Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer's disease screening and staging
Abstract
Introduction: Blood proteins are emerging as candidate biomarkers for Alzheimer's disease (AD). We systematically profiled the plasma proteome to identify novel AD blood biomarkers and develop a high-performance, blood-based test for AD.
Methods: We quantified 1160 plasma proteins in a Hong Kong Chinese cohort by high-throughput proximity extension assay and validated the results in an independent cohort. In subgroup analyses, plasma biomarkers for amyloid, tau, phosphorylated tau, and neurodegeneration were used as endophenotypes of AD.
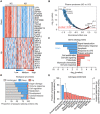
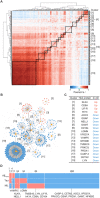
Results: We identified 429 proteins that were dysregulated in AD plasma. We selected 19 "hub proteins" representative of the AD plasma protein profile, which formed the basis of a scoring system that accurately classified clinical AD (area under the curve = 0.9690-0.9816) and associated endophenotypes. Moreover, specific hub proteins exhibit disease stage-dependent dysregulation, which can delineate AD stages.
Discussion: This study comprehensively profiled the AD plasma proteome and serves as a foundation for a high-performance, blood-based test for clinical AD screening and staging.
Keywords: Alzheimer's disease; biomarker panel; diagnosis; disease staging; neurodegenerative disease; plasma proteome; prognosis.
© 2021 The Authors. Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
H.Z. has served on scientific advisory boards for Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, and CogRx; has given lectures in symposia sponsored by Fujirebio, AlzeCure, and Biogen; and is a cofounder of Brain Biomarker Solutions (BBS) of Gothenburg AB, which is part of the GU Ventures Incubator Program (outside submitted work). All other authors declare no conflicts of interest.
Figures




References
-
- Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid‐β biomarkers for Alzheimer's disease. Nature. 2018;554:249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

