Detection of Tumor DNA in Human Plasma with a Functional PLL-Based Surface Layer and Plasmonic Biosensing
- PMID: 34032412
- PMCID: PMC8294610
- DOI: 10.1021/acssensors.1c00360
Detection of Tumor DNA in Human Plasma with a Functional PLL-Based Surface Layer and Plasmonic Biosensing
Abstract
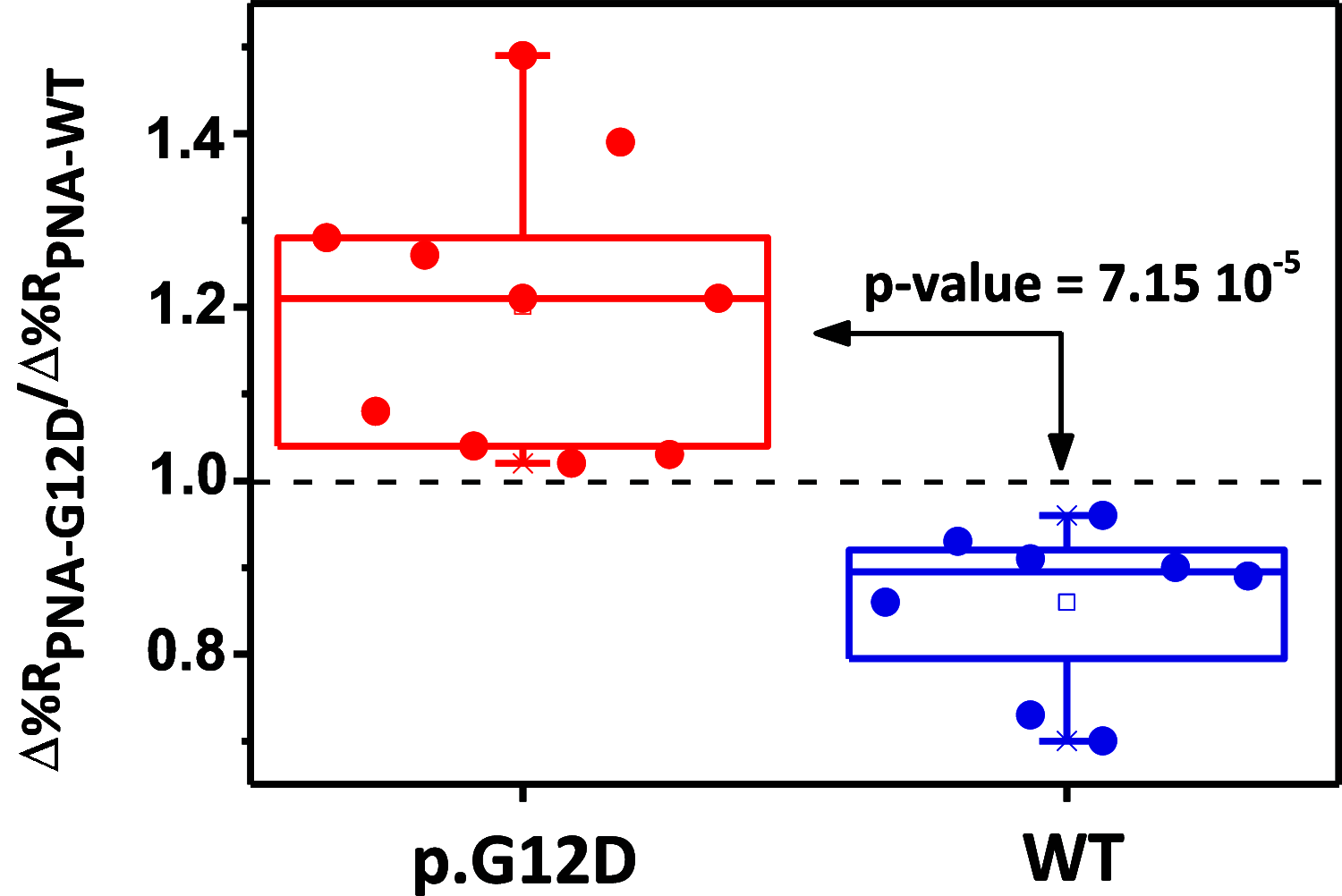
Standard protocols for the analysis of circulating tumor DNA (ctDNA) include the isolation of DNA from the patient's plasma and its amplification and analysis in buffered solutions. The application of such protocols is hampered by several factors, including the complexity and time-constrained preanalytical procedures, risks for sample contamination, extended analysis time, and assay costs. A recently introduced nanoparticle-enhanced surface plasmon resonance imaging-based assay has been shown to simplify procedures for the direct detection of tumor DNA in the patient's plasma, greatly simplifying the cumbersome preanalytical phase. To further simplify the protocol, a new dual-functional low-fouling poly-l-lysine (PLL)-based surface layer has been introduced that is described herein. The new PLL-based layer includes a densely immobilized CEEEEE oligopeptide to create a charge-balanced system preventing the nonspecific adsorption of plasma components on the sensor surface. The layer also comprises sparsely attached peptide nucleic acid probes complementary to the sequence of circulating DNA, e.g., the analyte that has to be captured in the plasma from cancer patients. We thoroughly investigated the contribution of each component of the dual-functional polymer to the antifouling properties of the surface layer. The low-fouling property of the new surface layer allowed us to detect wild-type and KRAS p.G12D-mutated DNA in human plasma at the attomolar level (∼2.5 aM) and KRAS p.G13D-mutated tumor DNA in liquid biopsy from a cancer patient with almost no preanalytical treatment of the patient's plasma, no need to isolate DNA from plasma, and without PCR amplification of the target sequence.
Keywords: cancer diagnosis; peptide nucleic acids; plasmonics; poly-l-lysine; surface plasmon resonance.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






Similar articles
-
Plasmonic aptasensor with antifouling dual-functional surface layer for lysozyme detection in food.Anal Chim Acta. 2023 Dec 1;1283:341979. doi: 10.1016/j.aca.2023.341979. Epub 2023 Oct 30. Anal Chim Acta. 2023. PMID: 37977796
-
A novel method for detecting genetic biomarkers in blood-based liquid biopsies using surface plasmon resonance imaging and magnetic beads shows promise in cancer diagnosis and monitoring.Talanta. 2025 May 1;286:127543. doi: 10.1016/j.talanta.2025.127543. Epub 2025 Jan 7. Talanta. 2025. PMID: 39798415
-
Low-fouling, mixed-charge poly-l-lysine polymers with anionic oligopeptide side-chains.J Mater Chem B. 2018 Dec 14;6(46):7662-7673. doi: 10.1039/c8tb01619d. Epub 2018 Nov 5. J Mater Chem B. 2018. PMID: 32254888
-
Design of peptide nucleic acid probes on plasmonic gold nanorods for detection of circulating tumor DNA point mutations.Biosens Bioelectron. 2019 Apr 1;130:236-244. doi: 10.1016/j.bios.2019.01.045. Epub 2019 Jan 29. Biosens Bioelectron. 2019. PMID: 30769288
-
A portable surface plasmon resonance sensor system for real-time monitoring of small to large analytes.J Ind Microbiol Biotechnol. 2005 Dec;32(11-12):669-74. doi: 10.1007/s10295-005-0044-5. Epub 2005 Nov 11. J Ind Microbiol Biotechnol. 2005. PMID: 16283397 Review.
Cited by
-
Design of Polymeric Surfaces as Platforms for Streamlined Cancer Diagnostics in Liquid Biopsies.Biosensors (Basel). 2023 Mar 18;13(3):400. doi: 10.3390/bios13030400. Biosensors (Basel). 2023. PMID: 36979612 Free PMC article. Review.
-
Interface Engineering of "Clickable" Organic Electrochemical Transistors toward Biosensing Devices.ACS Appl Mater Interfaces. 2023 Mar 1;15(8):10885-10896. doi: 10.1021/acsami.2c21493. Epub 2023 Feb 15. ACS Appl Mater Interfaces. 2023. PMID: 36791086 Free PMC article.
-
Sustainable Integration of Nanobiosensors in Biomedical and Civil Engineering: A Comprehensive Review.ACS Omega. 2025 Jun 10;10(24):25120-25157. doi: 10.1021/acsomega.5c00852. eCollection 2025 Jun 24. ACS Omega. 2025. PMID: 40584327 Free PMC article. Review.
-
Engineering of a DNA/γPNA Hybrid Nanoreporter for ctDNA Mutation Detection via γPNA Urinalysis.Adv Sci (Weinh). 2024 Sep;11(33):e2310225. doi: 10.1002/advs.202310225. Epub 2024 Jul 3. Adv Sci (Weinh). 2024. PMID: 38958527 Free PMC article.
-
Nanoparticle-Enhanced Surface Plasmon Resonance Imaging Enables the Ultrasensitive Detection of Non-Amplified Cell-Free Fetal DNA for Non-Invasive Prenatal Testing.Anal Chem. 2022 Jan 18;94(2):1118-1125. doi: 10.1021/acs.analchem.1c04196. Epub 2021 Dec 29. Anal Chem. 2022. PMID: 34964602 Free PMC article.
References
-
- Michael E. Taking Cancer out of Circulation. Nature 2020, 579, S6–S8. - PubMed
-
- Merker J. D.; Oxnard G. R.; Compton C.; Diehn M.; Hurley P.; Lazar A. J.; Lindeman N.; Lockwood C. M.; Rai A. J.; Schilsky R. L.; Tsimberidou A. M.; Vasalos P.; Billman B. L.; Oliver T. K.; Bruinooge S. S.; Hayes D. F.; Turner N. C. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. Arch. Pathol. Lab. Med. 2018, 142, 1242–1253. 10.5858/arpa.2018-0901-SA. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

