Protein-Ligand Binding Volume Determined from a Single 2D NMR Spectrum with Increasing Pressure
- PMID: 34032445
- PMCID: PMC8279561
- DOI: 10.1021/acs.jpcb.1c02917
Protein-Ligand Binding Volume Determined from a Single 2D NMR Spectrum with Increasing Pressure
Abstract
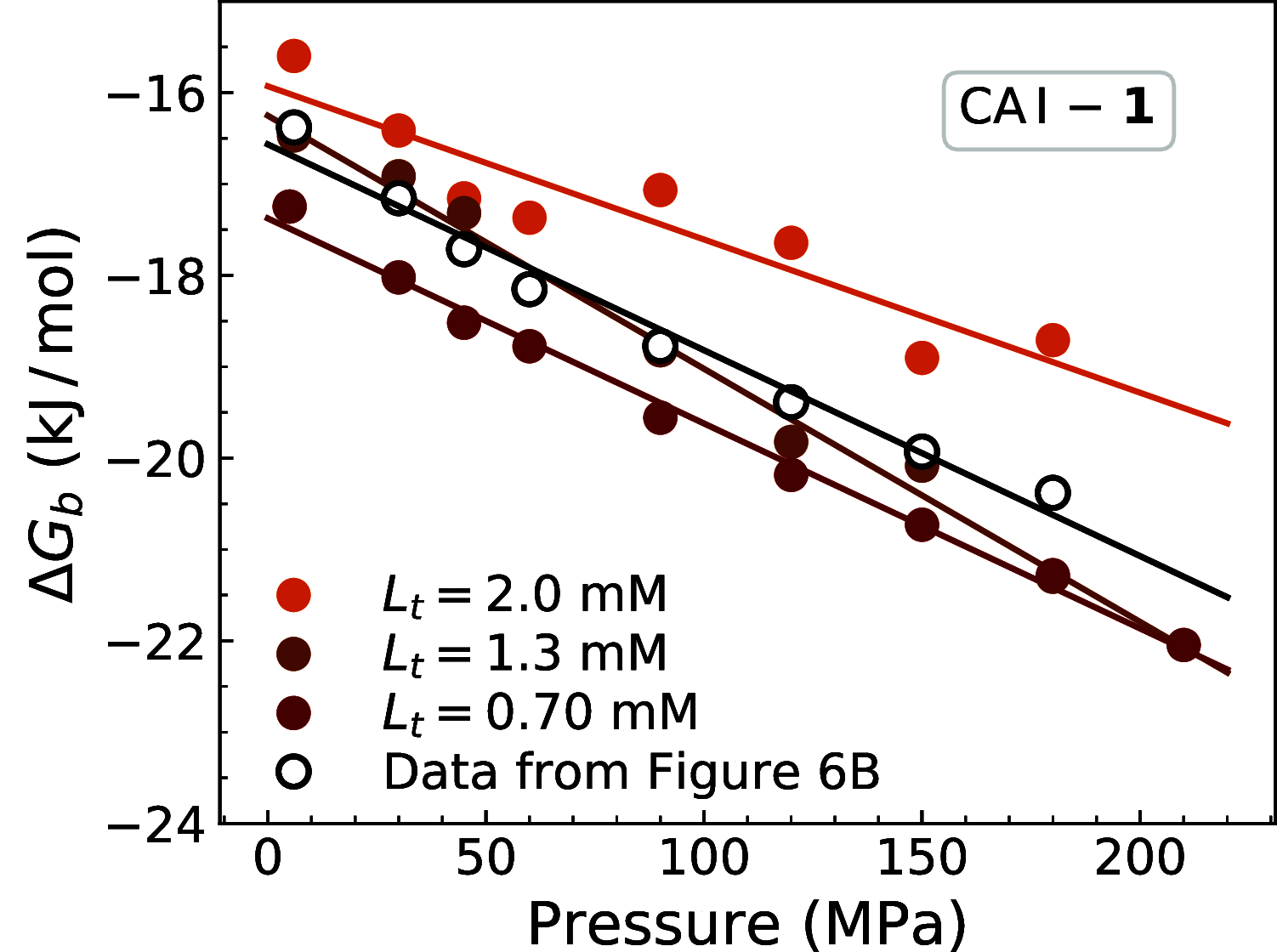
Proteins undergo changes in their partial volumes in numerous biological processes such as enzymatic catalysis, unfolding-refolding, and ligand binding. The change in the protein volume upon ligand binding-a parameter termed the protein-ligand binding volume-can be extensively studied by high-pressure NMR spectroscopy. In this study, we developed a method to determine the protein-ligand binding volume from a single two-dimensional (2D) 1H-15N heteronuclear single quantum coherence (HSQC) spectrum at different pressures, if the exchange between ligand-free and ligand-bound states of a protein is slow in the NMR time-scale. This approach required a significantly lower amount of protein and NMR time to determine the protein-ligand binding volume of two carbonic anhydrase isozymes upon binding their ligands. The proposed method can be used in other protein-ligand systems and expand the knowledge about protein volume changes upon small-molecule binding.
Conflict of interest statement
The authors declare no competing financial interest.
Figures









Similar articles
-
Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry.Biochemistry. 2004 Mar 9;43(9):2394-404. doi: 10.1021/bi035422a. Biochemistry. 2004. PMID: 14992576
-
Volumetric properties underlying ligand binding in a monomeric hemoglobin: a high-pressure NMR study.Biochim Biophys Acta. 2013 Sep;1834(9):1910-22. doi: 10.1016/j.bbapap.2013.04.016. Epub 2013 Apr 22. Biochim Biophys Acta. 2013. PMID: 23619242
-
Identifying ligands directly interacting with target protein in medicinal herbs by metabolomic analysis of T2-filtered HSQC spectra.J Pharm Biomed Anal. 2024 Sep 15;248:116329. doi: 10.1016/j.jpba.2024.116329. Epub 2024 Jun 27. J Pharm Biomed Anal. 2024. PMID: 38959759
-
Protein-Small Molecule Interactions by WaterLOGSY.Methods Enzymol. 2019;615:477-500. doi: 10.1016/bs.mie.2018.08.020. Epub 2018 Sep 20. Methods Enzymol. 2019. PMID: 30638539 Review.
-
Using chemical shift perturbation to characterise ligand binding.Prog Nucl Magn Reson Spectrosc. 2013 Aug;73:1-16. doi: 10.1016/j.pnmrs.2013.02.001. Epub 2013 Mar 21. Prog Nucl Magn Reson Spectrosc. 2013. PMID: 23962882 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

