Overexpressed DEPDC1B contributes to the progression of hepatocellular carcinoma by CDK1
- PMID: 34032605
- PMCID: PMC8436915
- DOI: 10.18632/aging.203016
Overexpressed DEPDC1B contributes to the progression of hepatocellular carcinoma by CDK1
Abstract
Background: Hepatocellular carcinoma (HCC) is the main type of primary liver cancer and shows a heavy burden worldwide. Its recurrence and mortality rate are still uncontrolled by the usage of present treatments. More attention has been focused on exploring specific genes that play important roles in HCC procession, and the function of DEP domain containing 1B (DEPDC1B) in HCC has not been researched.
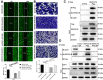
Methods: Immunohistochemical staining was used to detect the expression level of DEPDC1B in tumor tissues and adjacent normal tissues. After DEPDC1B and CDK1 knockdown in cell lines HEP3B2.1-7 and SK-HEP-1, MTT assay and colony formation assay was used to detect cell growth, flow cytometry assay was used to investigate cell apoptosis and cell cycle, wound-healing assay and Transwell assay were used to examine the tumor cell migration. Moreover, a xenograft model was constructed to research functions of DEPDC1B in tumor growth in vivo.
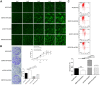
Results: The results show that DEPDC1B knockdown inhibit the progression of HCC, through inhibiting cell proliferation, migration, colony formation, leading to G2 phase arrest, and promoting cell apoptosis in vitro, and CDK1 was selected for further mechanic research according to the results of Human GeneChip prime view. The results of recovery experiment displayed that the functions of DEPDC1B on HCC progression were mediated by CDK1. DEPDC1B knockdown can also inhibit tumor growth in vivo.
Conclusions: The study confirmed that DEPDC1B knockdown restrains the tumor growth in vitro and vivo, and it can interact with CDK1 and rescued by CDK1. The study suggested that DEPDC1B was as a potential therapeutic target involved in HCC growth and progression.
Keywords: CDK1; DEPDC1B; hepatocellular carcinoma (HCC); migration; proliferation.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

